HTGAA: Genome Engineering
- John Glass (JCVI)
Class Material
- Class Slides
- Class Video
- Recitation Slides
- Recitation Video
- Related Readings & References
- Feedback Form
Grand Scale Genome Engineering of Viruses, Bacteria, and Eukaryotes
News of the COVID-19 pandemic dominates all media. There is lots of material about possible scientific solutions to help mitigate the disease. Many university and industrial scientific groups have shifted their attention to this new problem. Imagine that you are a scientist told by your advisor or boss to come up with a plan to make a difference. Possible ways to go about this include therapeutic drugs (small molecules like nucleoside analogs or biologicals like enzymes, antibodies, or nucleic acids), new vaccines, or new or better diagnostics. Here is your assignment:
- Describe your new product. Do this using 6 sentences or less.
- I used to work in the pharmaceutical industry on developing new therapeutics to treat infectious diseases. One thing that was drilled into us was that it takes 3000 days to get a new drug to market once it has been discovered. That’s about 8 years. During that time things like toxicology studies using animals, pharmacokinetics, and plans for large scale synthesis of the drugs are the first things determined. Then there are Phase !, II, and III clinical trials to do. Under ordinary circumstances, every day is precious because a new drug or vaccine patent life diminishes while all this research is going on. Now haste is even more important because we are in a pandemic, and people are dying. If you roam around the internet, you will see that there are ways to accelerate some of these processes. President Trump talks about having a vaccine in a few months. That seems farfetched, but as I mentioned in the lecture, Moderna is already testing a mRNA-based vaccine against the corona virus. Many companies are developing new diagnostic approaches. Provide a bullet point outline of the steps needed to get your product approved by the US FDA and into clinical use. Be brief, my goal here is that you gain some understanding of the regulatory processes for medical solutions to a problem like the current pandemic.
- How would your approval process be different if you were developing a product for a new rabies vaccine (i.e. something that was not urgent)? Try to write this in six sentences of less.
One of the most unexpected findings from the JCVI minimal bacterial cell project was that almost one third of the genes (149 of 473) encoded proteins of no known function.
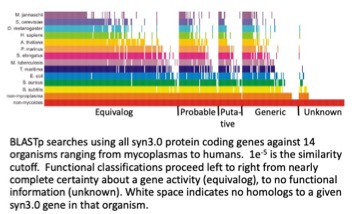
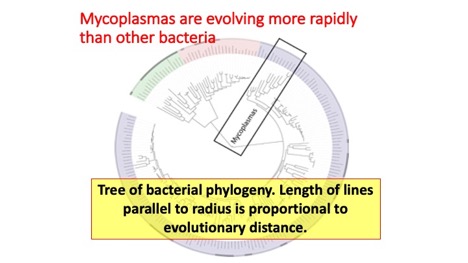
Why do so many of the essential genes have no known function? I offer a possible clue. Look at the dendogram below. It is based on 16S rRNA sequences. Make a hypothesis using this clue or offer an idea or ideas of your own.
As stated above, in the minimal cell JCVI-syn3.0, 149 of 473 genes made products of unknown function. That is about 31%. Would the predecessor of JCVI-syn3.0, wild type Mycoplasma mycoides have a higher percentage of unknown function genes? Why? How about for Escherichia coli, the most studied of all bacteria. Would it have a higher fraction of genes of unknown function? Why? Short answers to the why questions are acceptable.
In an age where scientists like us can design and build new organisms and viruses, what is acceptable and non-acceptable research? When Eckard Wimmer synthesized a poliovirus genome to produce virus from digital information or David Evans made horsepox virus with a synthetic genome there was a public outcry and widespread condemnation. The same happened when my JCVI team constructed a bacterium with a synthetic genome. Consider the HTGAA class with Megan Palmer and how it relates to what we know is possible and likely to be possible.
Related Readings & References
I offer more articles than you will be able to get through. They are organized by topic and more or less in the same order the topic are introduced in the lecture. Unless you are more motivated than I think necessary for the HTGAA class, I suggest you read just for concepts, not for experimental details.
Reverse Genetics of Viruses and Using Synthetic Biology to address COVID-19
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290(5498):1972-4. PubMed PMID: 11110665. This paper was the first synthesis of a complete viral genome and also a major paper for Hepatitis C Virus research. The synthesis of the 8001 bp HCV genome was only mentioned in the references and so hardly noticed. I recommend you just look for the synthetic method in the referenecs (#9).
- Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297(5583):1016-8. PMID: 12114528. This paper is often cited as the first synthesis of a complete viral genome. It got a lot of press because someone showed how a virus could be made from digital information. This raised a lot of fears about the dangers of synthetic biology, but no actions were taken.
- Smith HO, Hutchison CA, 3rd, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15440-5. PMID: 14657399. Previous viral genomes of about the same size took six months to two years to make. This effort by the JCVI made a synthetic genome in two weeks. This approach led to the JCVI synthesis of complete bacterial genomes.
- Morowitz HJ. The completeness of molecular biology. Israel journal of medical sciences. 1984;20(9):750-3. PMID: 6511349. This is a short, but amazing article that portends the construction of the JCVI minimal cell and discusses how this research will catalyze the expansion of our knowledge about cell biology.
- Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329(5987):52-6. Epub 2010/05/22. doi: 10.1126/science.1190719. PMID: 20488990. The JCVI team developed the methods to synthesize and boot up bacterial genomes. There were Science papers in 2007, 2008, and 2009 that described the methods developed. This paper presents the first cell with a chemically synthesized genome, JCVI-syn1.0. At the time this organism was described as a synthetic cell, but that is inaccurate.
- Hutchison CA, 3rd, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351(6280):aad6253. PMID: 27013737. This describes the design and construction of the first minimal bacterial cell, JCVI-syn3.0. It is acutally a near minimal bacterial cell. SInce 2016, the JCVI team has identified a number of JCV-syn3.0 genes that are not essential. Remarkably, of the 473 genes encoded by JCVI-syn3.0, in 2016 we did not know the function of 149 of those genes. Since then that number of genes of unknown function has dropped to 102.
- Ostrov N, Landon M, Guell M, Kuznetsov G, Teramoto J, Cervantes N, et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353(6301):819-22. PMID: 2754017. Harvard's George Church led a remarkable effort that produced an E. coli strain that uses only 57 codons. This genome reduced organism has the potential to radically alter our approaches for building proteins that use non-standard amino acids.
- Fredens J, Wang K, de la Torre D, Funke LFH, Robertson WE, Christova Y, et al. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019;569(7757):514-8.. PMID: 31092918. This organism, which was completed a few years after the Church lab's 57 condon E. coli appears to be a bit more robust. This and the previous reading show two strategies to produce bacteria to be used to produce proteins with non-standard amino acids.
- Pretorius IS, Boeke JD. Yeast 2.0-connecting the dots in the construction of the world's first functional synthetic eukaryotic genome. FEMS Yeast Res. 2018;18(4). PMID: 29648592. This is a summary of where the Yeast 2.0 project was and where it was going near the end of the endeavor. It is a good read.
- Brown DM, Chan YA, Desai PJ, Grzesik P, Oldfield LM, Vashee S, et al. Efficient size-independent chromosome delivery from yeast to cultured cell lines. Nucleic acids research. 2017;45(7):e50. PMID: 27980064. It is my belief that building synthetic chromosomes is the easy part of such efforts. Installing them in target cells so that they function as designed in more challenging. Here, my University of Maryland graduate student describes how he was able to improve delivery of synthetic chromomes that are cloned as yeast artifical chromosomes into cultured mammalian cells by a factor of 1000. This has implications for the Human Artificial Chromosome field.
- Brown DM, Glass JI. Technology used to build and transfer mammalian chromosomes. Exp Cell Res. 2020:111851. PMID: 31952951. This is a short review about synthetic human and other types of artificial chromosomes.
- From the GP-Write website I suggest you take a look at this website. It gives you an idea where GP-Write, the effort to make higher eukaryotes with synthetic genomes is going.
- Eisenstein M. How to build a genome. Nature. 2020;578(7796):633-5. PMID: 32094921. This is a quick overview of the science of building whole genomes for viruses, bacteria, and eukaryotes.