Week 11: Engineering the gut microbiome
Engineering the gut microbiome
The research I have chosen was published in 2016 by Han et al., where they cultivated microalgae with bacteria strains.
The bacteria Muricauda sp. were co-cultured with microalga Tetraselmis chuii, Cylindrotheca fusiformis and Nannochloropsis gaditana in extra organic carbon containing medium
To solve the media problem, the bacteria reduce the dissolved oxygen concentration and consume the organic material excreted by microalgae. In turn they produce biotin, cobalt amine and thiamine
Many microalgae can grow on light and chemical energy. Although it would be reasonable to think that the bacteria and microalgae will compete for nutrients if they’re grown in the same cultivation system, the competition can be abated by adding excessive nutrients or continuously adding nutrients.
.

Photo credit: Han et al., 2016
I would be curious to automate the selection of organism to co-culture. If for example we had a chip where we could culture different combinations of microbes (eg. a chip with 1,000 options), we would be able to select the more “symbiotic combinations” of 2,3 or even tenths of organisms
One of the great challenges in microbiology currently is culturing “unculturable” microbes. Propose a methodology for how you might explore this significant space of uncultured microbes.
I guess that most unculturable microbes are those that live in very specific conditions that we cannot replicate.
By giving them the media we think they need, we might be promoting the growth of certain microbes instead of others within the sample.
Therefore, playing with low concentration of certain substrates might be enough to allow the growth of those microbes while containing
the growth from the others. We might even be able to use a micromanipulator to separate the different cells and then do a high screening
on culture conditions for each of them.
Review an article on an artificial gut-on-a-chip technology. What scientific hypotheses are they testing with this in vitro tool? Could you propose an upgrade or innovation to their technique to enable the exploration of other scientific hypotheses? Provide an example of at least one hypothesis you would explore with your proposed system.
I have selected this article by Poceviciute et al, 2019
They created an oxygen gradient across the endothelium-epithelum-lumen axis to culture human endothelial and epithelial cells with a diverse microbiota
The chip has 2 channels separated by a permeable membrane. Endothelial cells grew in the lower channel and epithelial cells in the upper one, where they got in contact with the microbiota.
They co-cultured two established cell lines (human intestinal microvascular endothelial cells, and Caco2 epithelial cells) and a model anaerobe (Bacteroides fragilis) on the chip.
On the left hand side of the image are the hypothesis they were testing and on the right one, the follow-on hypothesis they want to test. Below is the chip design
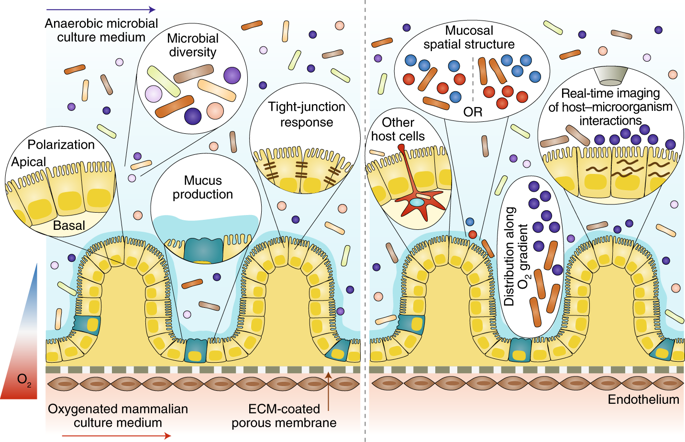
Photo credit: Poceviciute et al, 2019