HTGAA: 3D bioprinting and biofabrication
“hiPSC derived organoids as suitable models for regenerative medicine and drug screening”
The state of stem cell research has rapidly progressed over the last several years, especially with the advent of human induced pluripotent stem cells (hiPSCs), which have provided new insights into fundamental biology and allowed us to better understand human disorders and subsequent development of novel treatments. hiPSCs are remarkable in that they mimic human embryonic stem cells in their ability to differentiate into almost any cell type yet are generated from somatic cells (generally fibroblast skin cells) through a process termed cellular reprogramming (Fig. 1). During this section of the course we’ll be working with George Church’s own hiPSC line, the infamous PGP1 hiPSC line (generated from his own fibroblasts) with the aim of differentiating them into cardiac organoids, termed Engineered Heart Tissues (EHTs) (1). Organoids are 3D tissue-like structures comprised of multiple cell types which orginate from stem cells. They undergo self-organization and are able to mimic to a certain degree the architecture and functionality of in vivo organs.
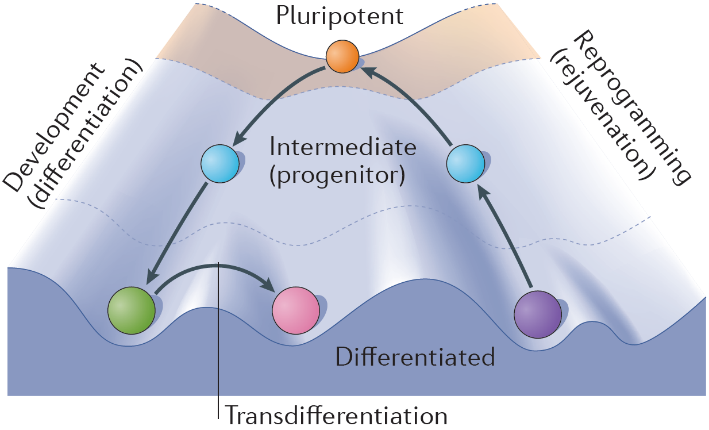
Given that the same factors that determine cell fate and function in vivo also determine the progression of cell differentiation in vitro, our ability to grow a 3-dimensional tissue (organoid) in the laboratory is therefore directly related to our ability, as engineers, to recapitulate the natural microenvironment. This principle is referred to as the biomimetic paradigm (i.e. “copying nature”). Hence, for tissue engineering applications, major efforts have been invested into characterizing native tissue environments, and describing these environments by parameters that may be recapitulated experimentally (e.g. the convection of blood through perfusion, the stiffness/porosity of scaffolds, the exposure of cells to a mechanical loading or electrical field stimulation, etc).
Our interest in cardiac organoids stems from the saddening statistic of cardiovascular diseases being the lead cause of death in the United States and across the world. The devasting economical and physical toll this inflicts on countries requires adequate model systems to further understand the etiology of these diseases and help work towards therapeutics and cures. More recently the advent of SARS-CoV2 has shown a proclivity towards cardiac injury and dysfunction, evident in 20-30% of hospitalized patients as well as increased rates of mortality in patients with pre-existing cardiovascular disease. Suitable model systems are required to recapitulate in vivo settings and properly assess the effects viruses like SARS-CoV2 have on organs like the heart. We will expose the PGP1 stem cell line to a stepwise cardiac induction protocol to generate the EHTs (1, 6).
Pre-homework questions:
- From an ethical and moral standpoint, list pros and cons of working with hiPSCs as opposed to embryonic stem cells?
- If we are to use organoid models as surrogates for in vivo tissues, what type of functional attributes would be critical to add/engineer into such organoid systems?
- Can you think of any limitations in current organoid and or organ on a chip systems in recapitulating human biology?
- If you were to theorize an experiment about improving current organoid models, which aspects of the microenvironment would you consider adding beyond chemical?
- In the tissue engineering paradigm, what properties are important to provide from a scaffold and bioreactor?
The homework component will include:
- Read prescribed articles and answer questions.
- Live streamed treatment of EHTs with student input of which drugs to use, reasoning behind selection of drugs (what do each of them do?), concentrations to use and if incremental dosing is required?
Experimental setup for EHT drug screening
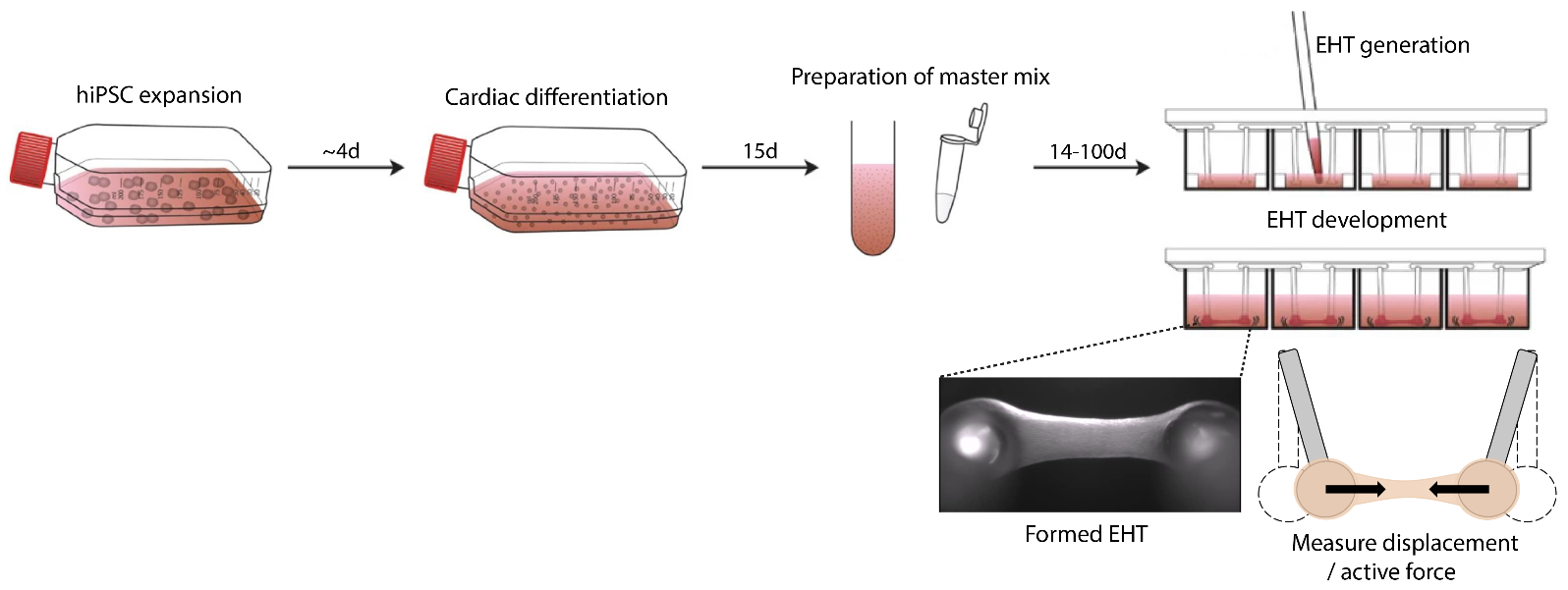
Cardiac organoids / EHTs will be generated following a well-established published protocol (1, 6). Briefly, hiPSCs are differentiated towards a mesodermal fate which subsequently transitions towards a cardiac lineage. The EHTs will be generated by mixing the hiPSC derived cardiomyocytes in a fibrin hydrogel and seeded onto silicone racks containing elastomeric poles so that the cardiomyocytes can attach and form contracting cardiac organoids (Fig. 2). We will utilize these EHT organoids for the treatment of selected drugs where you will subsequently generate your own custom analysis pipeline to calculate contractile force / displacement of the EHTs.
EHT drug screening homework component
We will live-stream this section for you to decide the experimental setup. The following drugs will be available to choose from:
- Caffeine
- Oxytocin
- Atosiban
- Isprenaline
- Theobromine
- Adrenaline
- Norephinephrine
Briefly hypothesize how each of these drugs would affect the heart and based off your research select the top drug you would like to test on the EHTs. Also discuss how you would select “appropriate” concentrations to work with and what is the standard used today in drug screening in determining drug concentrations for clinical settings (e.g. EC50).
Once we have treated the EHTs with the selected drugs, we will provide microscopy recordings where your objective is to analyse and calculate the displacement of the micropillars and ultimately active/contractile force using either ImageJ, Matlab, etc. We will cover this in more detail during the recitation. Conclude from your calculations if the compounds had worked as theorized and whether there are any experimental aspects you would alter after the fact. The objective is for students to work together and share their data from the various drugs to compare.
_______________________________________________________________________________
Extra section on Mycelium presented by Bob Hendrikx (TU Delft, Loop)
We have a guest presenter from TU Delft, Bob Hendrikx, presenting his fascinating work on mycelium (e.g. the living coffin: https://www.dezeen.com/2020/09/16/bob-hendrikx-living-cocoon-mycelium-coffin/). Please view the following TEDx talk (https://www.youtube.com/watch?v=XI5frPV58tY) and his website (www.bobhendrikx.com) to see some of the amazing work Bob is carrying out with mycelium. During the recitation Bob will walk us through his work as well as a fun at home component for the local Boston students.
References
1. Breckwoldt, K., Letuffe-Brenière, D., Mannhardt, I., Schulze, T., Ulmer, B., Werner, T., ... & Hansen, A. (2017). Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nature protocols, 12(6), 1177.
2. Tandon, N., Cimetta, E., Bhumiratana, S., Godier-Furnemont, A., Maidhof, R., & Vunjak-Novakovic, G. (2013). Bioreactors for tissue engineering. In Biomaterials Science (pp. 1178-1194). Academic Press.
3. Bhumiratana, S., Bernhard, J. C., Alfi, D. M., Yeager, K., Eton, R. E., Bova, J., ... & Vunjak-Novakovic, G. (2016). Tissue-engineered autologous grafts for facial bone reconstruction. Science translational medicine, 8(343), 343ra83-343ra83.
4. Tandon, N., Goh, B., Marsano, A., Chao, P. H. G., Montouri-Sorrentino, C., Gimble, J., & Vunjak-Novakovic, G. (2009, September). Alignment and elongation of human adipose-derived stem cells in response to direct-current electrical stimulation. In 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society (pp. 6517-6521). IEEE.
5. Tandon, N., Cannizzaro, C., Chao, P.H.G., Maidhof, R., Marsano, A., Au, H.T.H., Radisic, M. and Vunjak-Novakovic, G., 2009. Electrical stimulation systems for cardiac tissue engineering. Nature protocols, 4(2), p.155.
6. (Optional reading) Mills, R.J., Titmarsh, D.M., Koenig, X., Parker, B.L., Ryall, J.G., Quaife-Ryan, G.A., Voges, H.K., Hodson, M.P., Ferguson, C., Drowley, L. and Plowright, A.T., 2017. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proceedings of the National Academy of Sciences, 114(40), pp.E8372-E8381.
7. (Optional reading) Takahashi, K. and Yamanaka, S., 2016. A decade of transcription factor-mediated reprogramming to pluripotency. Nature reviews Molecular cell biology, 17(3), p.183.