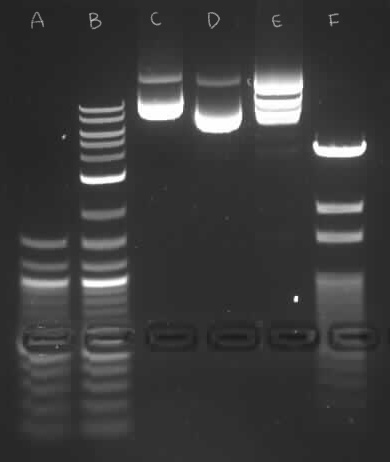
1. What software are you using to read the sequence files?
o Benchling
2. What are the distances between the PstI (CTGCAG) sites in each plasmid?
o pPSU1:
494, 1994, 994, 694, 794, 894, 4094
o pPSU2:
1494, 44, 94, 194, 294, 394, 494, 594, 4094
3. What are the distances between the EcoRV (GATATC) sites in
each plasmid?
o pPSU1:
494, 994, 1494, 1994, 4994.
o pPSU2:
744, 2994, 3994
o Add
6 bases to each distance (the size of the cut site)
4. Where do PstI and EcoRV cut within their recognition sequences?
o PstI: CTGCA|G
� G|ACGTC
o EcoRV: GAT|ATC
�
CTA|TAG
o Add
6 bases to each distance (the size of the cut site)
5. What gene is encoded into the plasmids?
o AmpR, for ampicillin resistance
6. How many copies of the plasmid would you roughly expect in each cell?
o Since they are high copy number plasmids, you would expect thousands of copies of the plasmids in each cell.
7. How do the plasmids' replication relate to that of pBR322
(genbank)?
o pBR322 is a medium copy number plasmid, having about 10-1000 copies in each cell, so pPSU1 and pPSU2 replicates more in cells.
Pro Challenge
Add a restriction site into the E.
coli genome through lambda-red recombination.
For this week, provide the oligo design that
achieves your proposed genomic edit and describe what impact this change may
have on the genetic code. In later weeks, you will carry out this recombination
within the electroporator you build, and soon after measure
your genome editing efficiency. Recall, George described lambda red recombination as being so
precise that it can do what CRISPR can only wish to accomplish, for now.
I will try to introduce a restriction site for
the enzyme EcoRV, with the cut site GATATC, in the
lacZ gene of E. coli. There currently exists an EcoRV
cut site at location 1126 of the lacZ gene. I would like to introduce a site at
location 1492. Currently, the bases at location 1492-1497 are GATATT, so a
point mutation of T1497C would turn this into an EcoRV
cut site, and could be done with a ssDNA oligo.
However, it would have the consequence of interrupting the lacZ gene and making
it nonfunctional.
Forward primer:
GCCCGGTGCAGTATGAAGGCGGCGGAGCCGACACCACGGCCACCGATATCatgagtattcaacatttccg
Uppercase: lacZ sequence, ending at base 1496. Bold: point mutation. Lowercase: AmpR beginning.
Reverse primer:
CAGCCGGGAAGGGCTGGTCTTCATCCACGCGCGCGTACATCGGGCAAATAttaccaatgcttaatcagtg
Uppercase: lacZ sequence reverse complement, ending at base 1498. Lowercase: AmpR end reverse complement.