Step 1: Digest the pUC19 plasmid with the restriction enzyme PvuII to generate the linear blunt-ended backbone fragment.
Step 2: Amplify PCR fragments for the gene assembly of amilCP mutants.

The first and second steps of this assignment was fairly straight and forward. By this time we are fairly familiar with the way digestion and fragments can interact to cause bacterial changes once they are prepared correctly.
Step 3. Purify the DNA products from your reactions using the Zymo DNA Clean & Concentrator™-25 Kit.
The following protocol is copied from Zymo Research and based on silica adsorption.
For each reaction: In a 1.5 ml microcentrifuge tube, add 5x volumes of DNA Binding Buffer to each volume of reaction.
Mix briefly by vortexing.Transfer each mixture into a separate Zymo-Spin™ Column in a Collection Tube.Centrifuge for 30 seconds at 13,000 rpm (~17,900 x g).
Discard the flow-through.Add 200 µl DNA Wash Buffer to the column. Centrifuge at for 30 seconds at 13,000 rpm (~17,900 x g). Repeat the wash step.
Transfer the column to a new 1.5 ml microcentrifuge tube and add 25 µl of DNase/RNase-free water directly to the column matrix.
Let sit at room temperature for one minute.Centrifuge for 30 seconds at 13,000 rpm (~17,900 x g) to elute.

Step 5. Setup your Gibson Assembly reaction in PCR tubes
Clean stage with Kimwipe soaked with DI water.
Wipe stage with dry Kimwipe.Add 2 uL DI water on stage and make blank measurement.
Wipe stage with dry Kimwipe.For each DNA sample, add 2 uL and measure and record the concentration (ng/uL).
Wipe stage dry with Kimwipe between measurements.
Step 6. Heat shock assemblies into chemically competent E coli (stored across the hall in the -80C).
Move Gibson assembly into a cold block on ice.
Thaw chemically competent cells on ice for 10 minutes.
Aliquot 50 ul cells into a PCR strip kept in a cold block on ice.Add 5 µl of the assembled product to the competent cells. Mix gently by pipetting up and down or by flicking the tube 4–5 times. Do not vortex.
Place the mixture on ice for 20 minutes. Do not mix.
Heat shock in the thermocycler at 42C for 30 seconds. Do not mix.
Transfer tubes to ice for 2 minutes.Combine heat-shocked cells with 500 ul of room-temperature SOC media in a new 1.5 ml microcentrifuge tube.
Spread 250 µl of the cells onto the ampicillin selection plates.
Incubate overnight at 37C in the warm room.
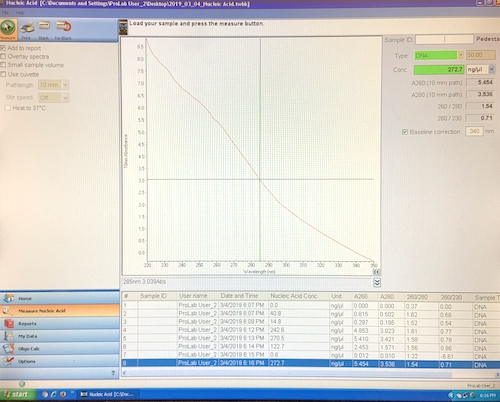
Step 4. Measure the concentration of your pPSU1 and pPSU2 DNA preps using the Nanodrop spectrophotometer with the following steps.
Clean stage with Kimwipe soaked with DI water.
Wipe stage with dry Kimwipe.Add 2 uL DI water on stage and make blank measurement.
Wipe stage with dry Kimwipe.For each DNA sample, add 2 uL and measure and record the concentration (ng/uL).
Wipe stage dry with Kimwipe between measurements.
My concentration measurements were 122.7, 272.7 and 68.5 (digest).
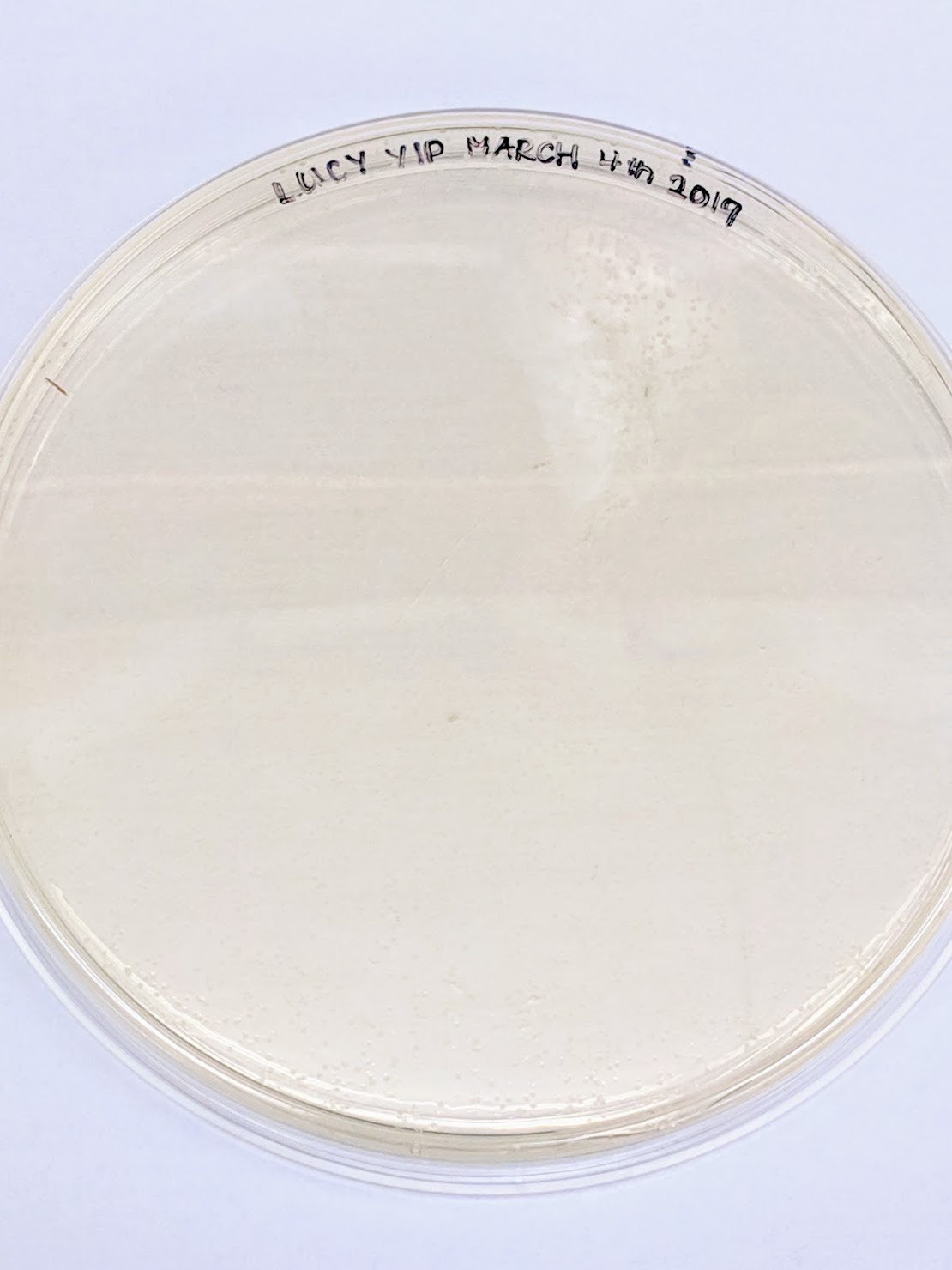
This was the outcome of the final place. There were a lot of growth present on the plate yet no colors, which could either be the issue with the Gibson / Plasmid or the amilCP. Since we are cloning by restriction digest, maybe most of the transformants are empty backbone (?) or the reaction was simply miss-calculated?