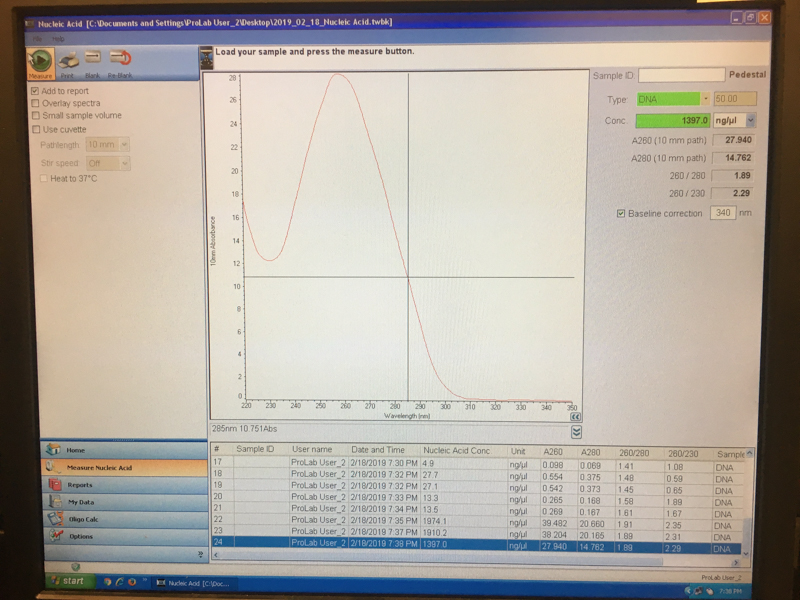
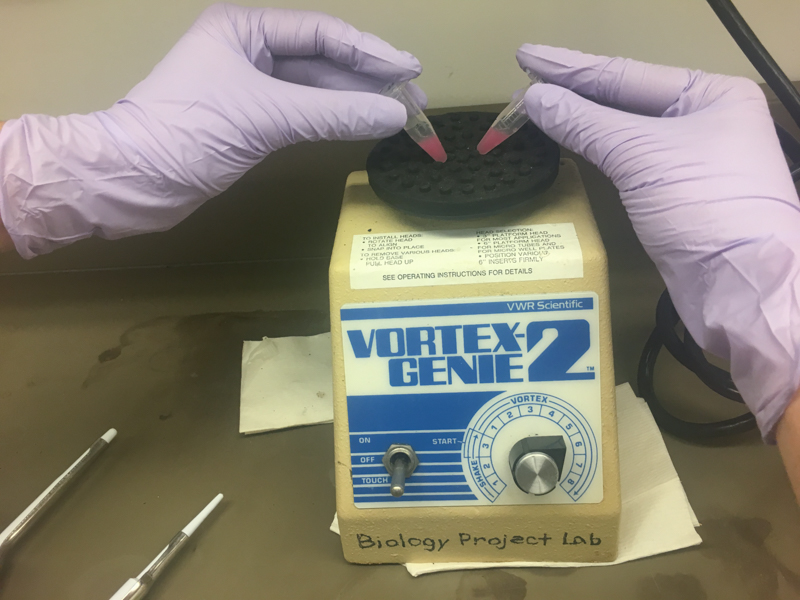
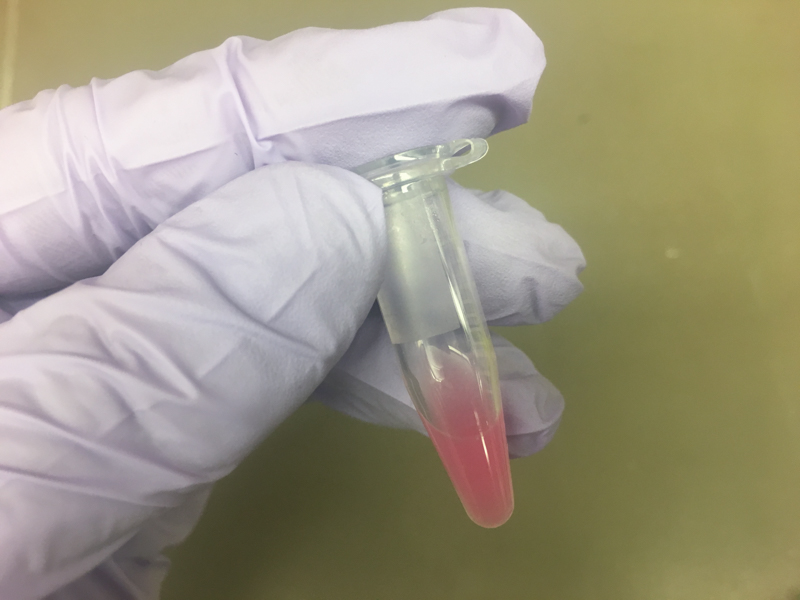
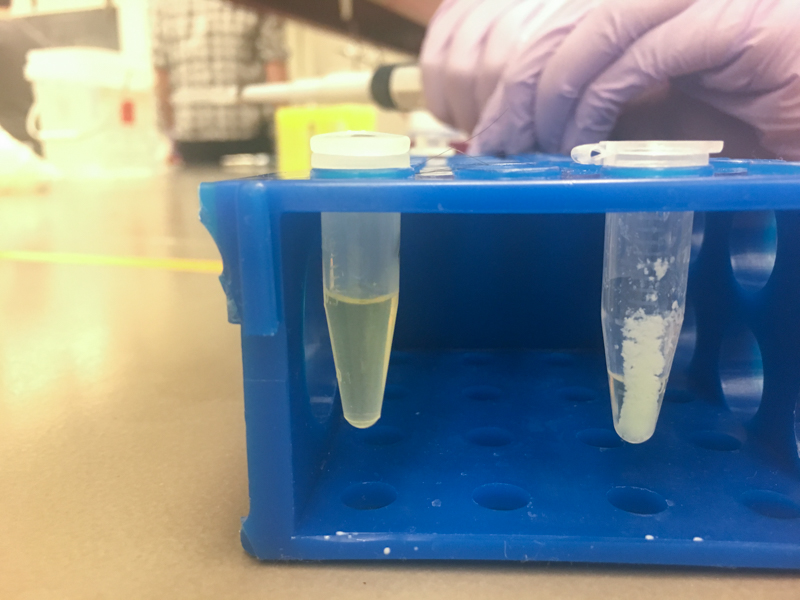
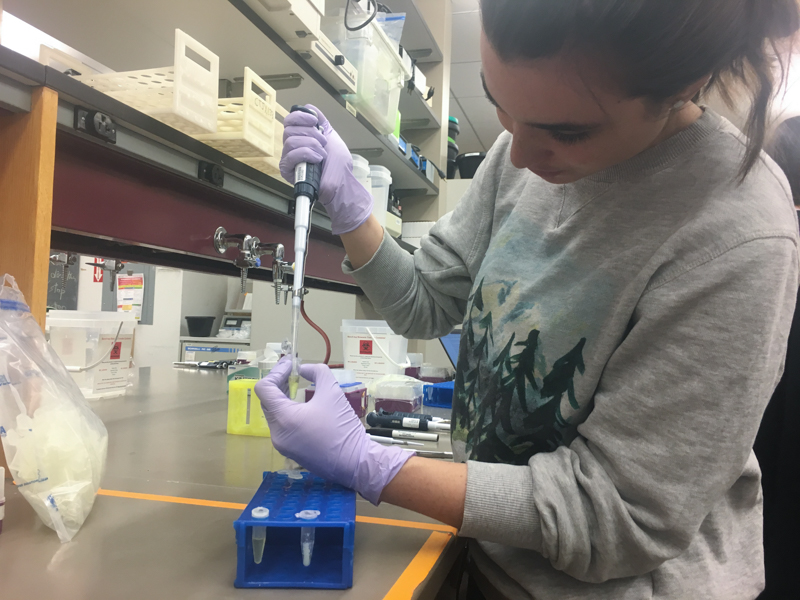
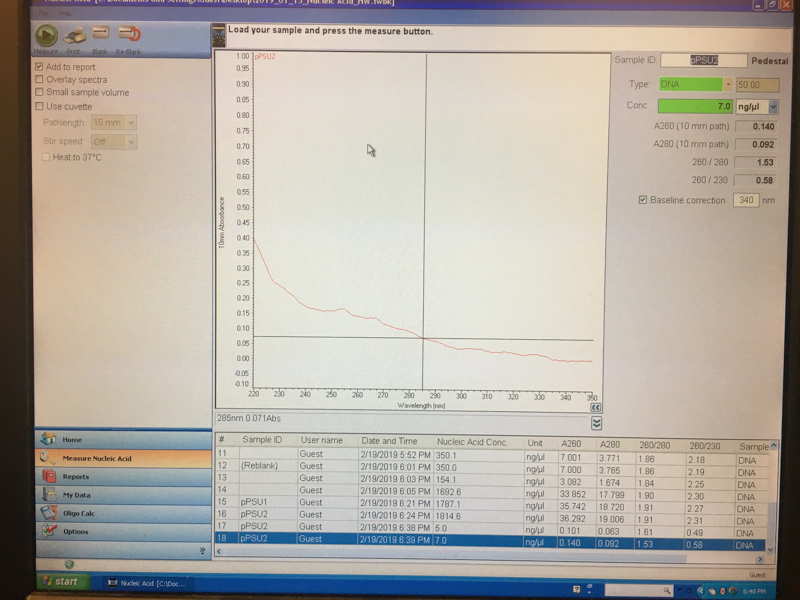
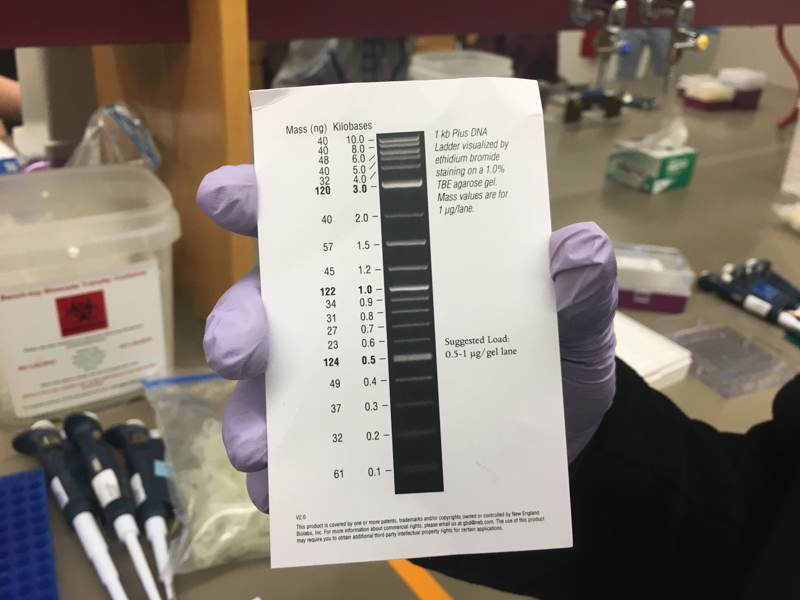
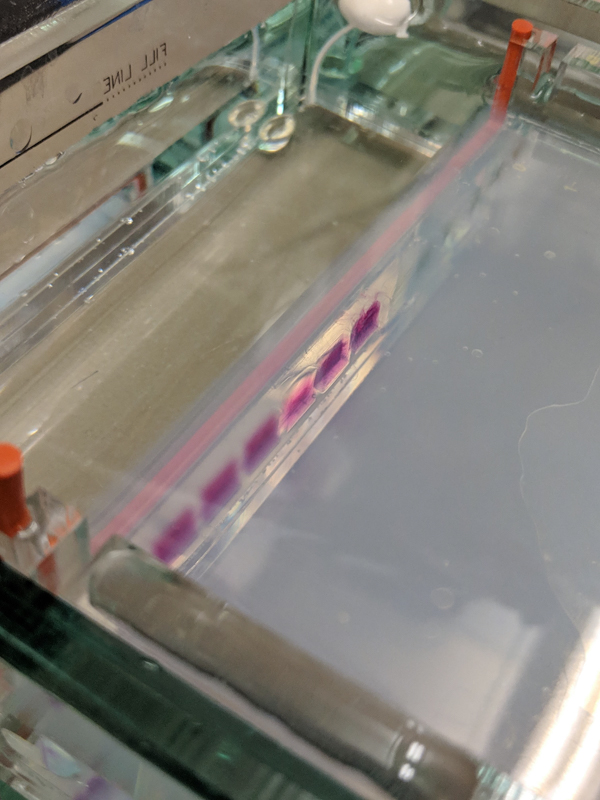
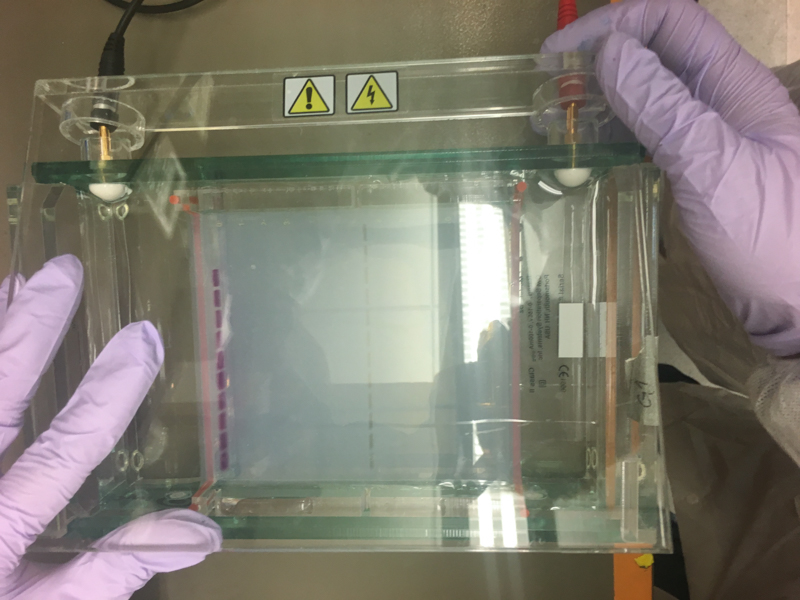
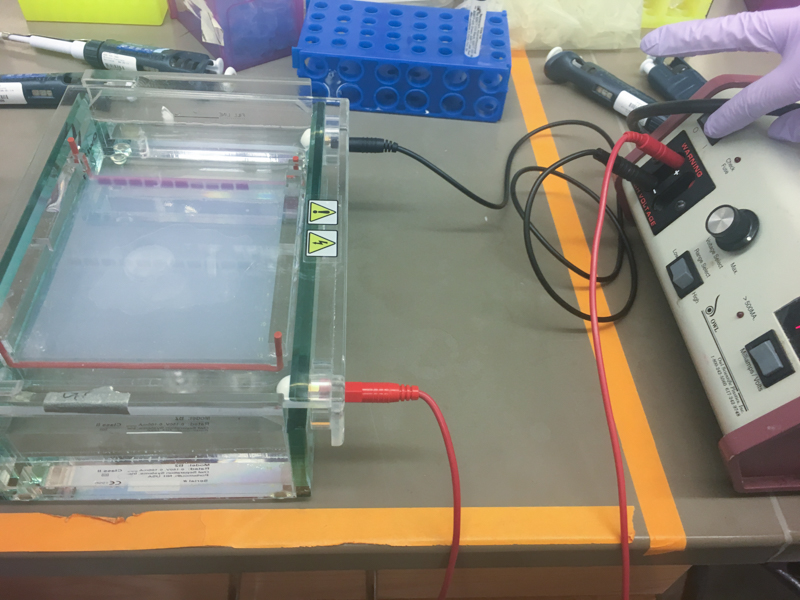
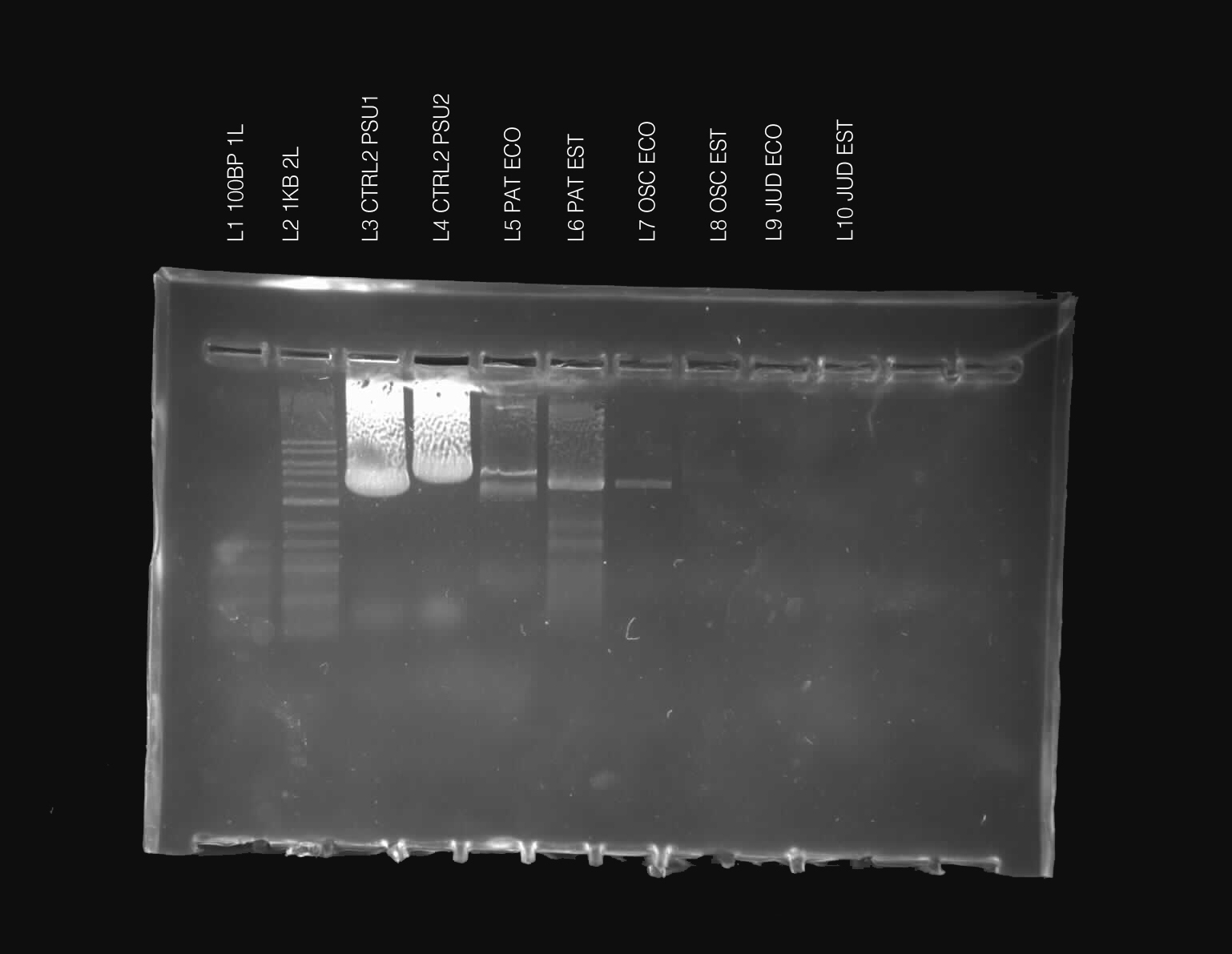
We will be working with Addgene plasmids pPSU1 and pPSU2 deposited by the Tan lab at Penn State University. In a 2017 paper, Henrici et al demonstrated how their plasmid designs can produce cheap DNA ladder. Note, DNA ladders are important references when measuring the size and topology of DNA products after an enzymatic reaction. The enzymes we will use for our example lab are restriction enzymes, which cut DNA at sequence-specific sites. Restriction enzymes are among the natural defense systems of bacteria to protect against viral bacteriophages. They were harnessed in the 1970's as a foundational technology for mapping and recombining DNA across all forms of life. Taking part in their characterization, George published a 1978 paper that sequenced the plasmid pBR322 to help determine the recognition sequence for the restriction enzyme AvaII. (From the experiment protocol)