Genome Engineering
Week 12
Genome Engineerings

Background of this Experiment (HTTGA)
http://fab.cba.mit.edu/classes/S66.19/S66.19/assignments/genomeengineering.htmlThe CRISPR-Cas9 system has been successfully harnessed for various genome editing applications, which have a wide range of implications in fields such as medicine, agriculture, bioenergy, and food security. CRISPR systems contain two components: a guide RNA (gRNA) and a CRISPR-associated (Cas) endonuclease. The gRNA is a short synthetic RNA composed of a scaffold sequence necessary for Cas-binding and a user-defined ∼20 nucleotide spacer that defines the genomic target to be modified. Thus, one can change the genomic target of the Cas protein by simply changing the target sequence present in the gRNA. To access specific targets, however, these enzymes require a protospacer adjacent motif (PAM), which is determined by DNA-protein interactions, to immediately follow the DNA sequence specified by the gRNA. The PAM sequence is essential for target binding, but the exact sequence depends on which Cas protein you use. For example, the standardly used Cas9 derived from Streptococcus pyogenes bacteria (SpCas9), for example, requires an 5'-NGG-3’ motif.
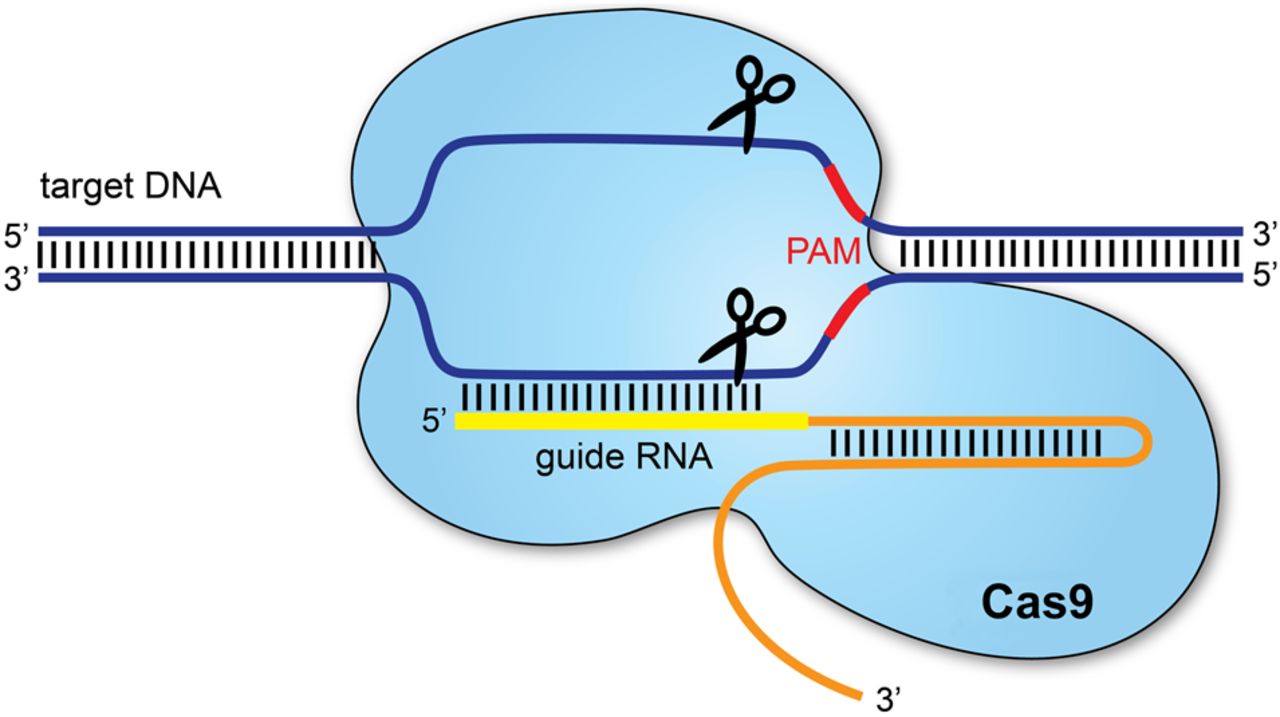
In this experiment, we evaluate the PAM binding of novel Cas9 enzymes through bacterial screen based on green fluorescent protein (GFP) expression conditioned on PAM binding. We performed the experiment by pipetting different Cas9 enzymes into separate bacteria cultures.
Beyond the experiment, we also have a very provocative lecture on Synthetic Genome by John Glass from J. Craig Venter Institute. Below are the thought exercise questions from lecture
- 1) What tasks must a cell perform to live?
- Ans : mutation and reproduction (so the population could evolve)
- 2) What bacterial innate immune mechanisms must be overcome to perform genome transplantation?
- Ans : CRISPR, RNAi
- 3) Can you suggest an alternative to genome transplantation to achieve genome scale engineering of bacterial genomes?
- Ans : 1) Invivo self-assemble of synthetic genome 2) Modification of the existing genome into target genome
- 3) What mammalian innate immune mechanisms must be overcome to efficiently install large DNA molecules in mammalian cells?
- Ans : Epigenetics Regulation, Splicing