Biodesign & DNA Experiment
Week 2
Biodesign

Background of this Experiment (HTTGA)
http://fab.cba.mit.edu/classes/S66.19/S66.19/assignments/biodesign.htmlWe will be working with Addgene plasmids pPSU1 and pPSU2 deposited by the Tan lab at Penn State University. In a 2017 paper, Henrici et al demonstrated how their plasmid designs can produce cheap DNA ladder. Note, DNA ladders are important references when measuring the size and topology of DNA products after an enzymatic reaction. The enzymes we will use for our example lab are restriction enzymes, which cut DNA at sequence-specific sites. Restriction enzymes are among the natural defense systems of bacteria to protect against viral bacteriophages. They were harnessed in the 1970's as a foundational technology for mapping and recombining DNA across all forms of life. Taking part in their characterization, George published a 1978 paper that sequenced the plasmid pBR322 to help determine the recognition sequence for the restriction enzyme AvaII.
In this experiment, we use Addgene plasmids pPSU1 and pPSU2 to create a DIY DNA ladder. Using two enzymes PstI (CTGCAG) and EcoRV (GATATC), it is possible to cut the two plasmid into smaller parts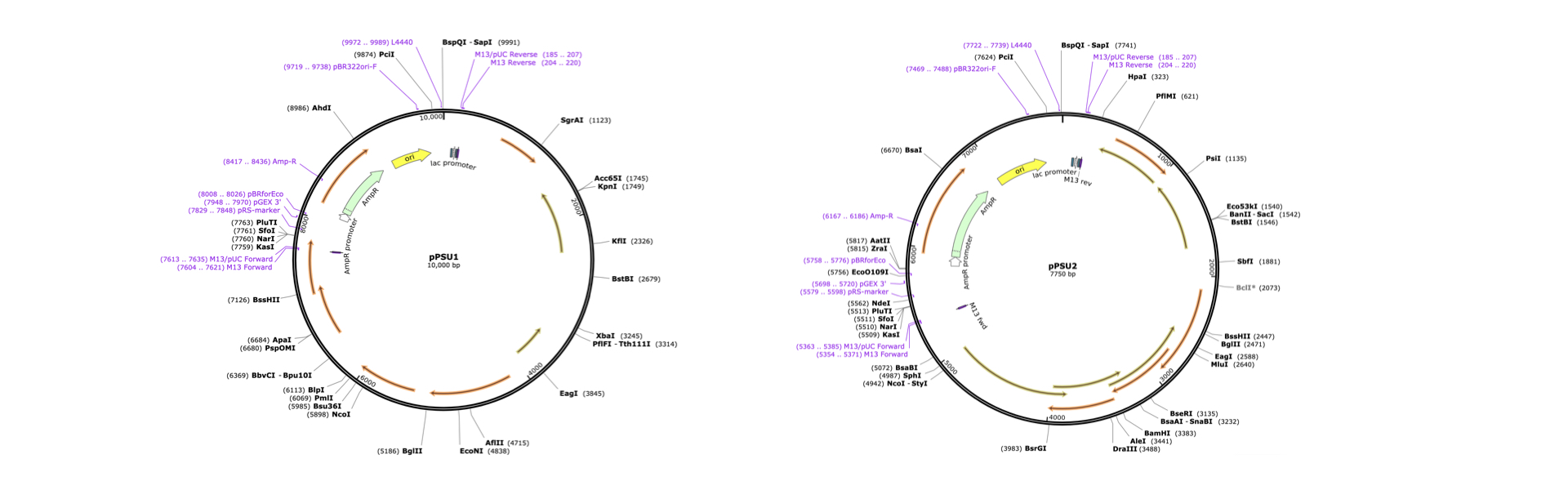
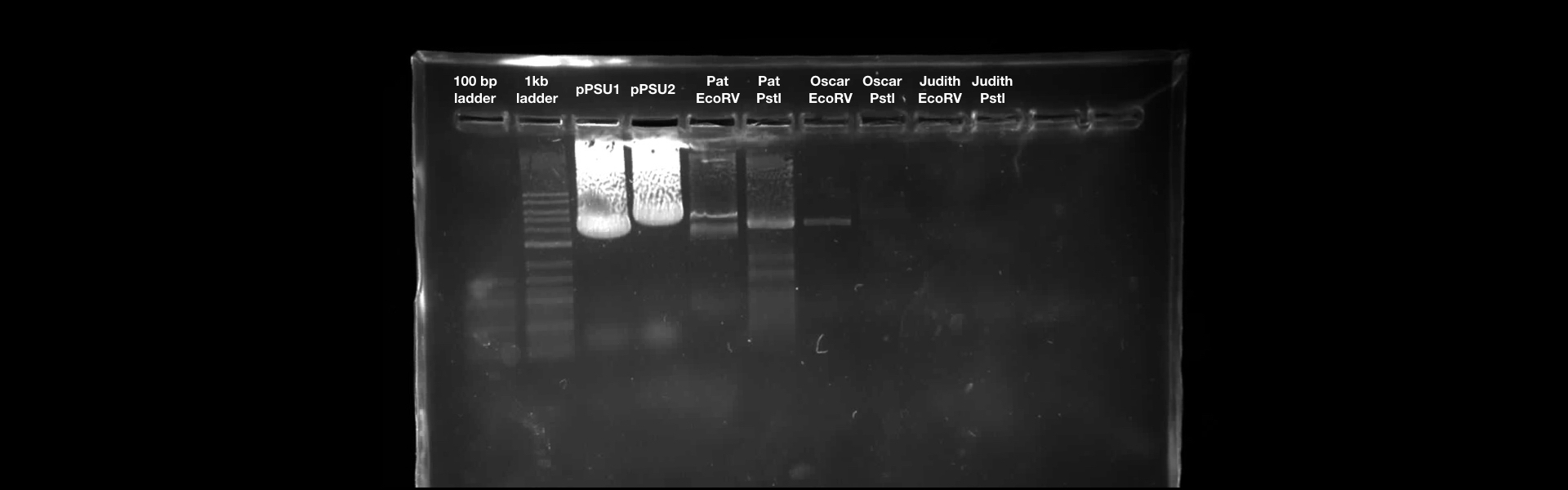
- The length of DNA from pPSU1 digested by PstI : 1696, 2196, 4196, 5196, 5896, 6696, 7596
- The length of DNA from pPSU1 digested by EcoRV : 241, 741, 1741, 3241, 5241
- The length of DNA from pPSU2 digested by PstI : 331, 1831, 1881, 1981, 2181, 2481, 2881, 3381, 3981
- The length of DNA from pPSU2 digested by EcoRV : 241, 991, 3991
We harvest the plasmid from E. coli culture using the ZymoPure Plasmid Miniprep Kit and elute with water.
We measured the concentration of our pPSU1 and pPSU2 DNA using the Nanodrop spectrophotometer. The concentration that we got for both sample are above 1500 ng/uL

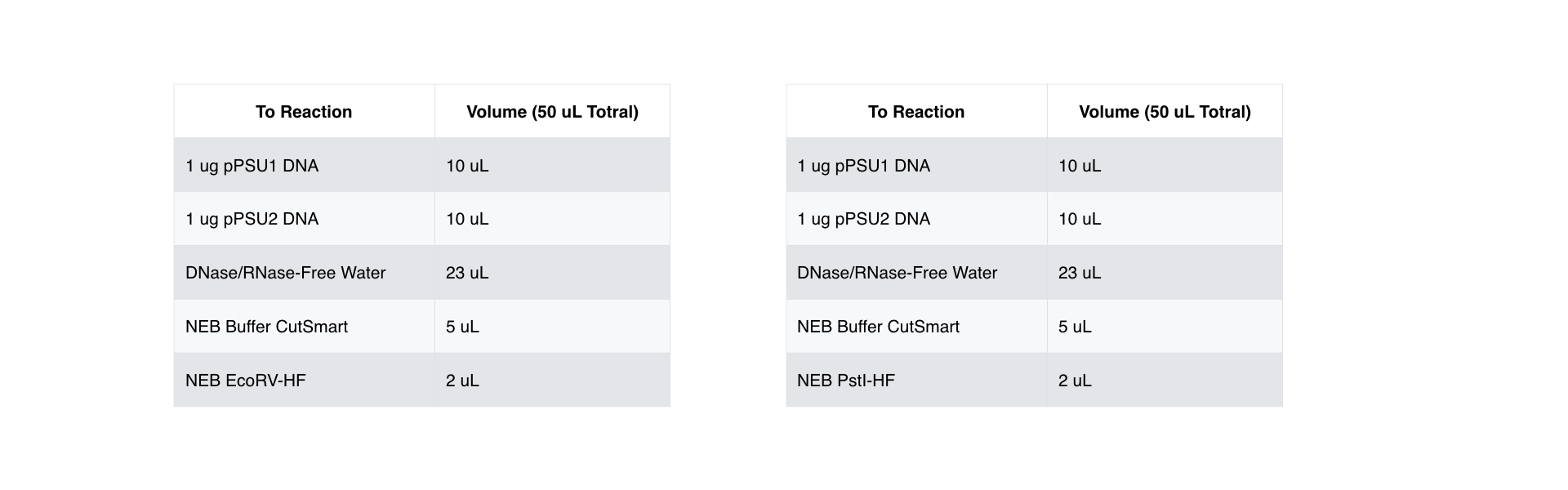
We diluted the plasmid to 100 ng/uL and total volume greater of 100 uL, and prepared the digestion reactions with PstI-HF and EcoRV-HF on both pPSU1 and pPSU2 DNA. Below is the reaction table
We prepared gel electrophoresis for visualizing our ladder.
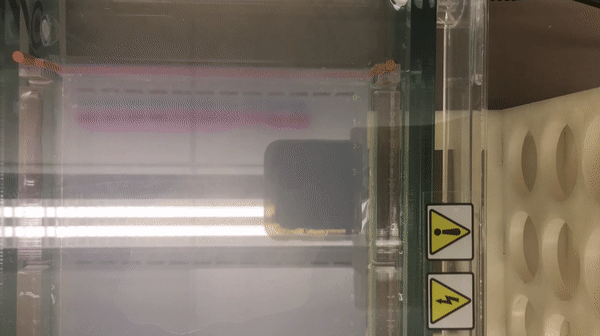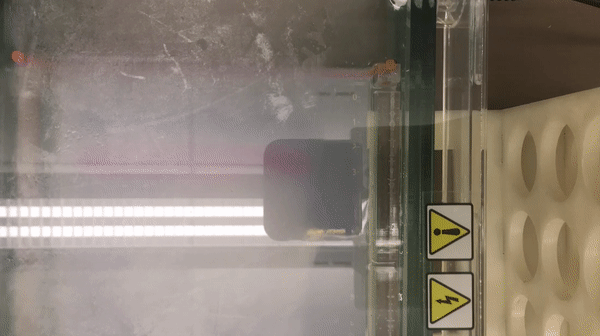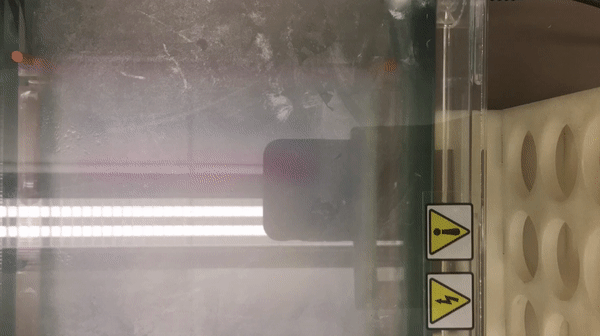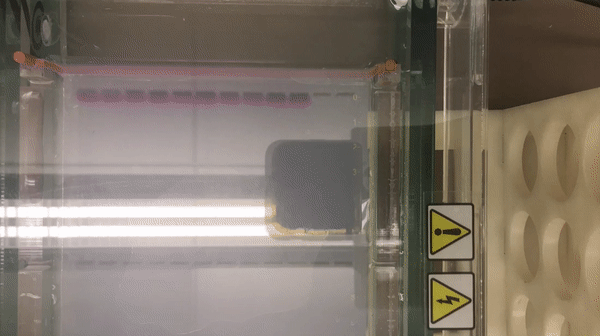

According to the result, we found that there is a high concentration of contaminants in the extracted DNA sample (lane 2 and 3). We believe that it might be RNA (as they also bind with the SYBR Safe). This contaminants might inhibit the performance of enzymes in our reaction. Thus, results in the incomplete cut on both EcoRV and Pstl reactions. This contaminants might result from the error in the DNA preparation process.