Biodesign: Creating a DNA Ladder
February 21, 2019
In this week's lab, we created a DNA ladder by using 2 restriction enzymes-EcoRV and PstI-on the plasmids PSU1 and PSU2, as discussed in Henrici et al. 2017. These plasmids were designed to have a variety of sizes of DNA when cut with EcoRV or PstI, ranging from 50 bp to 5000 bp.
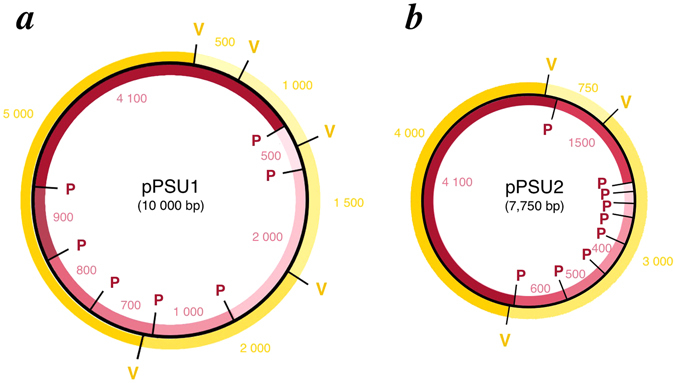
To visualize the plasmids, I used Benchling, an online DNA sequence visualization software. It enabled me to produce the two virtual digests as shown below, as a theoretical comparison for our experimental results.
Based on the above figures and a more detailed look at the sequences, the distances between the PstI and EcoRV sites on each plasmid are as follows:
1. PSU1 cut by PstI: 500 bp, 700 bp, 800 bp, 900 bp, 1.0 kb, 2.0 kb, 4.1 kb
2. PSU2 cut by PstI: 100 bp, 200 bp, 300 bp, 400 bp, 500 bp, 600 bp, 1.5 kb, 4.1 kb
PSU1 cut by EcoRV: 500 bp, 1.0 kb, 1.5 kb, 2.0 kb, 5.0 kb
PSU2 cut by EcoRV: 750 bp, 3.0 kb, 4.0 kb
The only gene on these plasmids is the ampicillin resistance gene, which was likely there to isolate the cells containing the plasmid by using ampicillin to kill cells without the plasmid. As a pUC9 derived plasmid, it has a high copy number, and it is likely each cell will have 500 to 700 plasmids. The pUC9 ori is derived from the pBR322 plasmid, but removes the rop gene that limits copy number and contains a mutation. Without these changes, the plasmid would have only had a copy number of 15 to 20 per cell.
