Synthetic Minimal Cells: Simplifying Biology
April 11, 2019
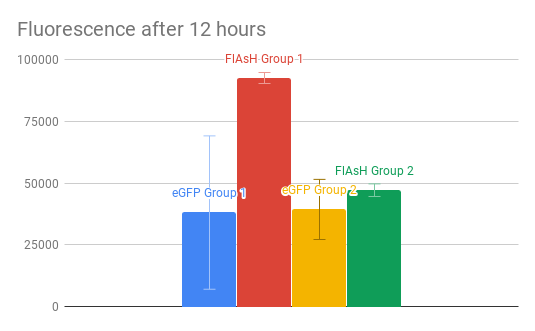
For the experimental portion of the assignment, we used cell-free Tx/Tl to produce GFP and FlAsH. The math to set up the reaction is here. More generally, eGFP is 239 aa long, whereas the FlAsH peptide is 1 kD, which implies it is approximately 9 aa long. This means the FlAsH peptide is significantly shorter, so it should be easier to transcribe more copies of it. E. coli has a transcription rate of 45 nucleotides/s and a translation rate of 15 aa/s. The quantum yield of eGFP is 60% and of FlAsH is 49%, meaning eGFP will produce more fluorescence than FlAsH will per mol. Based on the amino acid lengths, there will be 26.6 times as much FlAsH peptide produced as eGFP, but about 21.7 times as fluorescent because of the quantum yield difference. Assuming the binding of the FlAsH ligand and peptide is quick, we expect to see fluorescence in FlAsH first. The excitation of eGFP is 490 nm and emission is 510 nm, as seen in the graph below. The excitation of FlAsH is 500 nm and emission is 540 nm, as seen in the graph below.
However, these were not the results we got from experiment. Oddly, we were not able to see a difference in fluorescence from the blank tubes with our original wavelengths, and so we used 470 nm excitation and 515 nm emission to see results. We did get more FlAsH fluorescence than eGFP fluorescence, but not by quite as big a margin as expected. We also had oddly high background fluorescence. We attribute at least some of the oddity to poor maintainance of the RNAse free environment. There is also oddly high standard deviation on the eGFP and a relatively small standard deviation on FlAsH (though the two groups did get very different results for FlAsH).
For designing my synthetic cell, I would create a cell that would produce penicillin efficiently. This pathway would require alpha-aminoadipic acid and cysteine to begin, and would require a production method for alpha-aminoadipic acid. The output would be penicillin. This pathway could likely be realized without encapsulation, but the encapulsation provides a layer of protection for the penicillin. A genetically modified natural cell could do this, and has already in the mass production of penicillin, but it may be even more efficient to do it without growing an entire fungi. The outcome of this pathway would be the mass production of penicillin.
The membrane would likely be made of the phospholipids, cholesterol, a transport protein for alpha-aminoadipic acid, and a porin protein for penicillin. It would encapsulate the penicillin gene cluster, a cell free Tx/Tl system. The Tx/Tl system should be able to come from bacteria, as penicillin only interferes with the peptidoglycan cell wall and not bacterial Tx/Tl. It will import alpha-aminoadipic acid via transport proteins and output penicillin via its porins.
The enzymes and proteins necessary would be the tx/tl system, the OmpF porin from E. coli (shown to transport penicillin), and the transport protein for alpha-aminoadipic acid from rat astrocytes. The genes necessary would be the penicillin gene cluster pcbAB, pcbC, and penDE. Other necessary components would be cholesterol and alpha-aminoadipic acid. The function of the system could be measured by testing the potency of the antibiotic function of the extract of the system.