Week 2: BioDesign
Making a DNA ladder
In silico
I use Snapgene, which is free for MIT students, to view and manipulate nucleotide (and protein) sequences. In the pPSU1 plasmid, the PstI-cut fragments are 500, 700, 800, 900, 1000, 2000, and 4100 bp long; the EcoRV-cut fragments are 500, 1000, 1500, 2000, and 5000 bp long. In the pPSU2 plasmid, the PstI-cut fragments are 50, 100, 200, 300, 400, 500, 600, 1500, and 4100 bp long ; the EcoRV-cut fragments are 750 and 7000 bp long. Both plasmids contain the gene that encodes for a protein that grants resistance to the antibiotic ampicillin. Both plasmids, according to Addgene, have a high copy number, so we'd expect a few hundred copies/cell. This indicates much more efficient replication than the pBR322 plasmid which has 10-100 copies/cell, according to NEB. There are likely two reasons for this: 1) the pPSU plasmids lack the rop gene that pBR322 has - according to Bitesize Bio, the Rop protein negatively regulates plasmid replication; 2) the pBR322 has two antibiotic resistances genes comapred to pPSU's one, which results in a higher metabolic burden on the bacteria.
Miniprep
Learned some things that I had never thought to think about before:
- The column matrix comprises a silica gel membrane which electrostatically binds DNA
- The binding buffer and wash buffers must all be of the appropriate pH and ion concentration to ensure the DNA does not dissociate from the silica
Digestion
I deviated from the protocol slightly here: instead of adjusting the miniprepped DNA to 100ng/ul and using 10ul in the digestion reaction, I simply diluted ten-fold (total volume of 20ul, since the pipette has a lower limit of 2ul) and used a volume containing 1ug in the reaction. Makes the math a bit more straightforward for me.
Gel electrophoresis
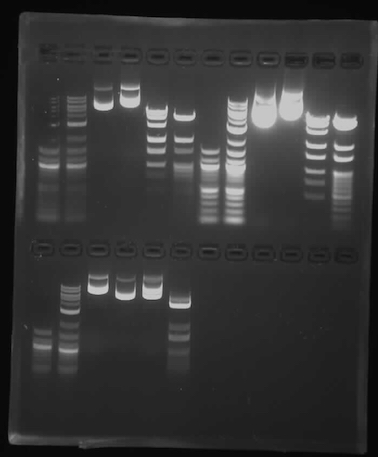
Since we used the smaller gel box with the small combs, we followed the instructions in mixing the samples for loading into the gel, but only loaded 30ul of sample, which seemed to be about the capacity of the wells we had.
The first six lanes below are mine, with the samples in the same order as is written in the class protocol. The bands are reasonably clean, and the enzyme-digested samples align with the reference ladders quite well. Interestingly, the middle two samples, which are undigested plasmid DNA, have multiple bands. Comparing to the reference ladder (src), the highest band in each lane is likely the intact plasmid, since plasmids travel more slowly in gel than linear DNA of the same length. It is odd that the left sample (pPSU1) traveled further, considering that it is 10kb compared to the right sample (pPSU2)'s 7.5kb. Possibly they got switched up. The lower bands in each lane are probably DNA that have been digested somehow, or at least linearized.