What software are you using to read the sequence files?
benching
What are the distances between the PstI (CTGCAG) sites in each plasmid?
pPSU1 494, 1994, 994, 694, 794, 894, 4094
pPSU2 1494, 44, 94, 194, 294, 394, 494, 594, 4094
What are the distances between the EcoRV (GATATC) sites in each plasmid?
pPSU1 494, 994, 1494, 1994, 4994
pPSU2 744, 2994, 3994
Where do PstI and EcoRV cut within their recognition sequences?
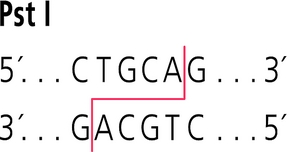

What gene is encoded into the plasmids?
AmpR gene encoded, ampicillin resistance
How many copies of the plasmid would you roughly expect in each cell?
high number copy plasmid ~1000
How do the plasmids' replication relate to that of pBR322 (genbank)?
pBR322 medium number 10-100 lower replication
FIRST WE ADDED ZYMOPURE P1 P2 P3 TO THE E.COLI CONTAINING PLASMID DNA WE WANT, pPSU1 AND pPSU2.
THE SOLUTIONS CUTS DNA AND REBINDS JUST THE PLASMID DNAS.


VORTEXING

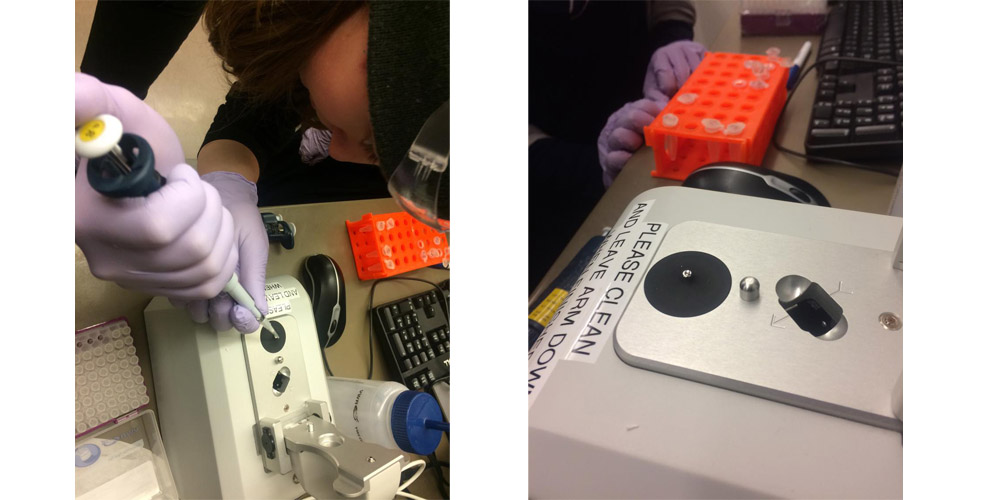
HONING OUR PIPETTING SKILLS

INCUBATING NEUTRALIZED LYSATE ON ICE

CENTRIFUGING

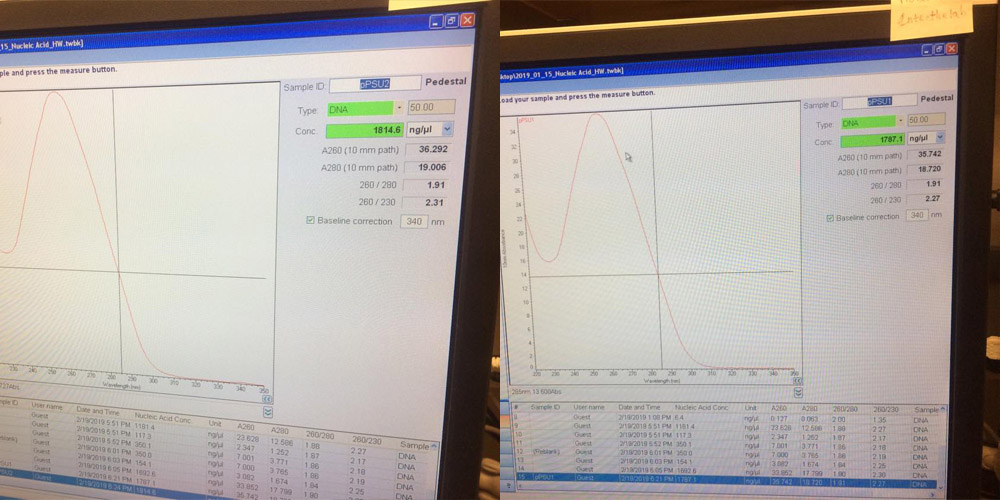
THEN WE ADDED A BINDING BUFFER AND TRANSFERED INTO COLUMN, FILTERED AND WASHED 3X.NEXT WE MEASURED THE CONCENTRATION OF OUR PSUs WITH A NANODROP MACHINE.
GOT A HIGH CONCENTRATION
WE DILUTED THE SOLUTION, ADDED ENZYMES TO CUT PLASMID DNA INTO SEVERAL LENGTHS.
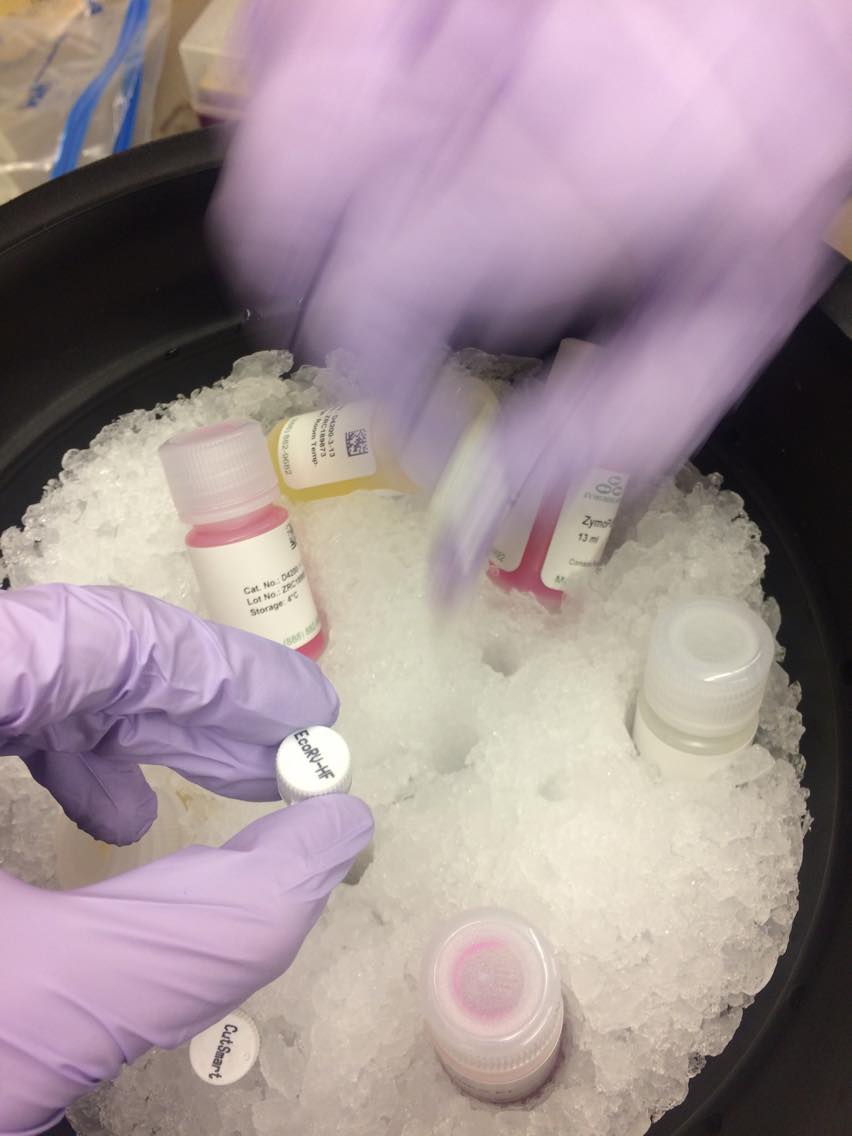
INCUBATED AT 37C TO LET ENZYMES WORK.

MEANWHILE RAHMA EXPLAINED THE BIOLOGY BEHIND IT TO US ON HER GLOVES
