What are the Gibson overlap sequences in our DNA assembly design? How is an overlap created between the cut pUC19 and the PCR amplified parts of the amilCP gene? See assignment lecture slides
PUC19 ORANGE AND BLUE REGIONS CAGCTG GTCGAC
What is the melting temperature for each overlap sequence and how do they compare to the incubation temperature for the Gibson assembly reaction? See http://biotools.nubic.northwestern.edu/OligoCalc.html
50C FOR GIBSON MIX pUC19_PvuII_LrgEnd1: 53.8C
pUC19_PvuII_LrgEnd2: 53.0C
What is the purpose of each enzyme in the Gibson assembly mix?
EXONUCLEASE CREATES 3' OVERHANGS, PLOYMERASE EXTENDS 3' ENDS FILLING IN GAP, LIGASE SEALS REMAINING MIX
1. Digest the pUC19 plasmid with the restriction enzyme PvuII to generate the linear blunt-ended backbone fragment.
2. Amplify PCR fragments for the gene assembly of amilCP mutants.

3. Purify the DNA products from your reactions using the Zymo DNA Clean & Concentrator™-25 Kit.

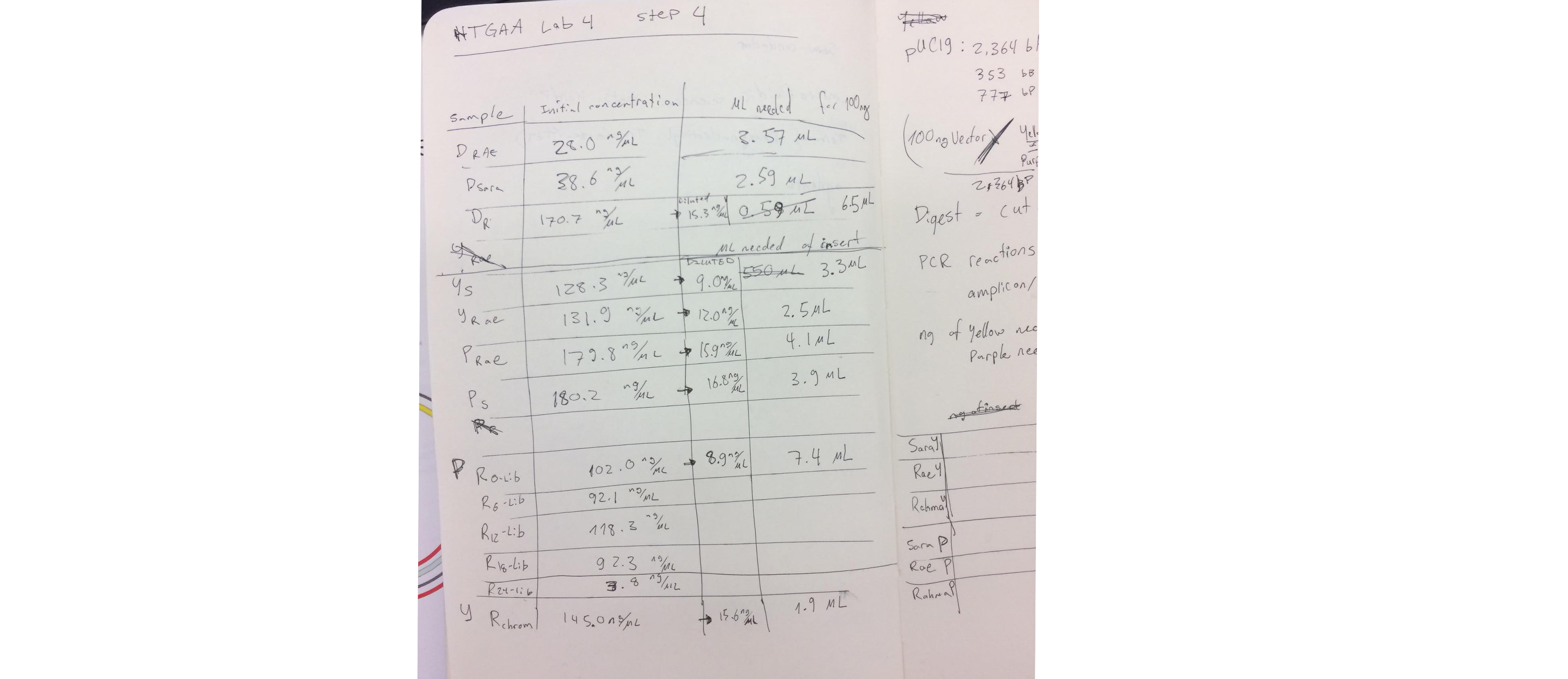
4. Measure the concentration of your purified DNA using the Nanodrop spectrophotometer.
5. Setup your Gibson Assembly reaction in PCR tubes.
6. Heat shock assemblies into chemically competent E coli (stored across the hall in the -80C).


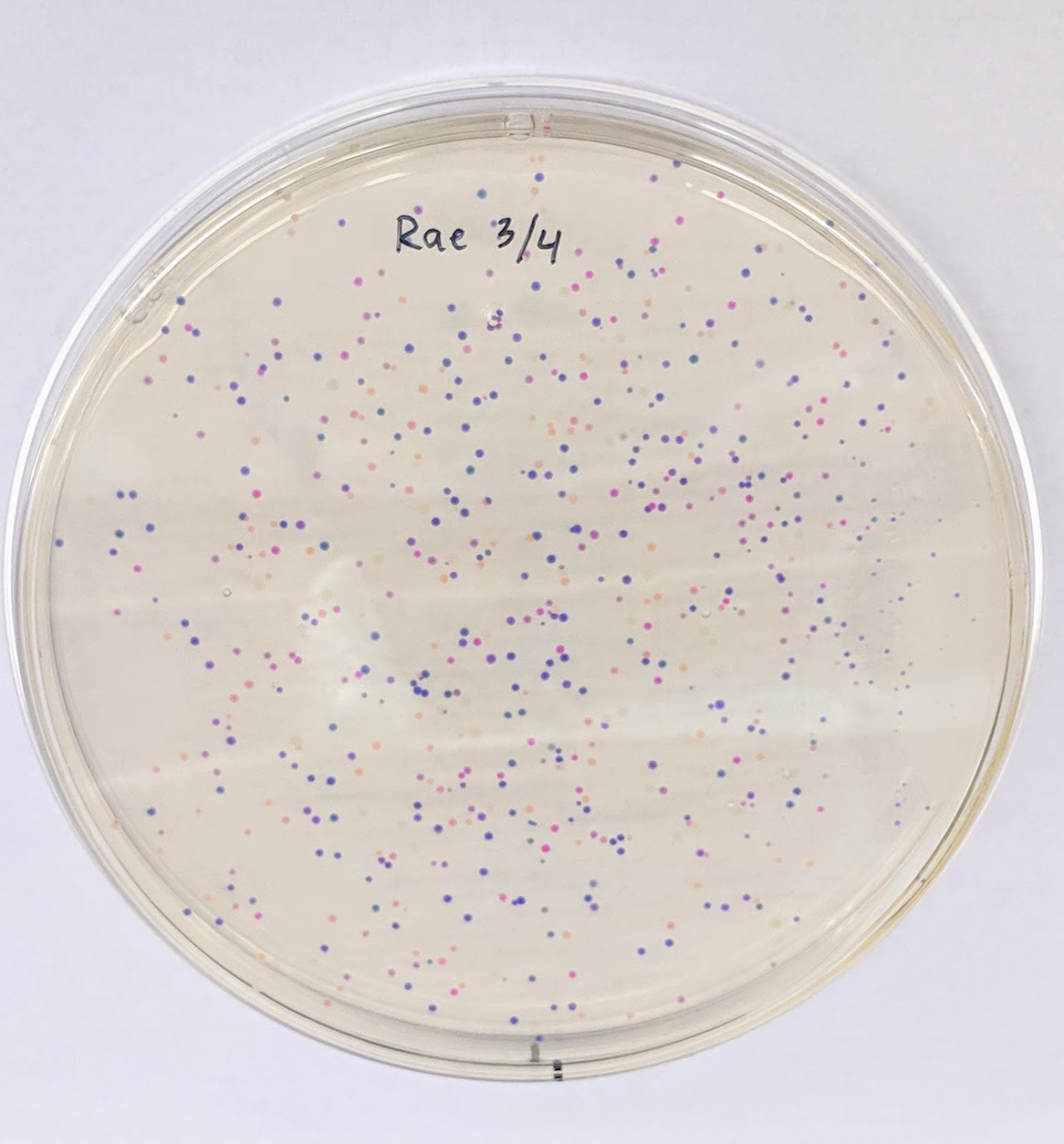
RESULTS