week 1: laser cutting
For laser cutting a press-fit construction kit, I thought it could be amusing to make a carboard version of the 'legos of life', as it were: DNA. Of course, DNA origami is a real a serious field of research in which people use Watson-Crick base pairing to tailor DNA structure on the nanoscale. My version, I suppose, is a slightly more macro DNA origami. (An aside: after I independently had this idea, a quick google search revealed I was not the first person who consider this. but since they charge $40 for a kit, I thought it would still be amusing to design my own version ab inito). DNA is a polymer of nucleotides, each of which contains a deoxyribose sugar, a nucleobase, and a phosphate. The sugars and phosphates provide the "backbone" of a DNA strand, and the nucleobases - which come in 4 varieties, 2 purines (adenine, guanine), and 2 pyrimidines (cytosine, thymine) - provide the substrate for information storage. Nucleobases bond with pairwise specificity (i.e. Waston-Crick bonding) due to hydrogen bonding (G and C form 3 h-bonds, A and 2, T).
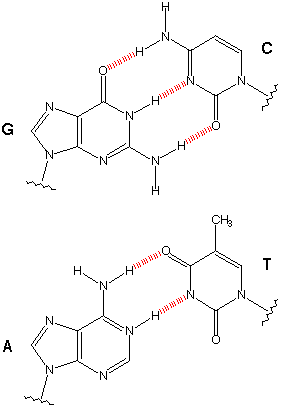
watson-crick bonding specificity
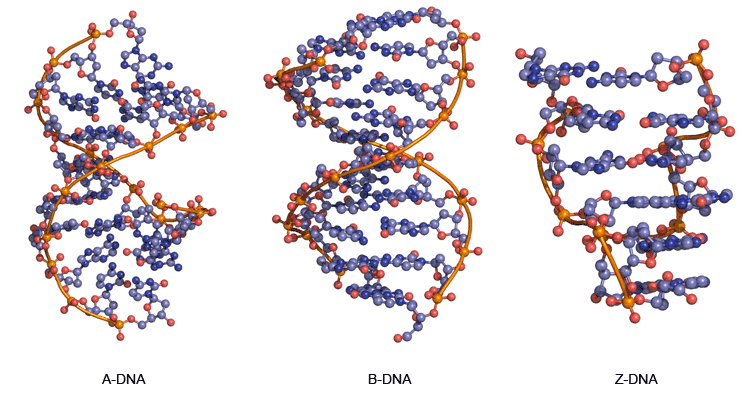
variants of 3D double helix structure in DNA
For my press-fit construction kit, I designed DNA components that could be assembled into a double helix molecule. Consistent with their chemistry, I designed pyrimidines as hexagons, and purines as pentagon-hexagon conjugates. Watson-crick specificity was encoded by the size of the slot designed for "hydrogen bonding" - G-C bonds allow for 2 widths of cardboard, and A-T only one (proper fidelity would require 3/2 bonds respectively, but this was more economical). The antimony design was parameterized via the cardboard thickness t, the helicity angle phi (setting the base pairs per turn), and the backbone inclination angle theta (setting the length per turn).

antimony rendering of DNA pieces CW, from top left: guanine, 2 h-bonds, 2 sugar-phosphate backbone units, cytosine

antimony rendering of 1 assembled base pair (A-T)

laser cutting DNA pices
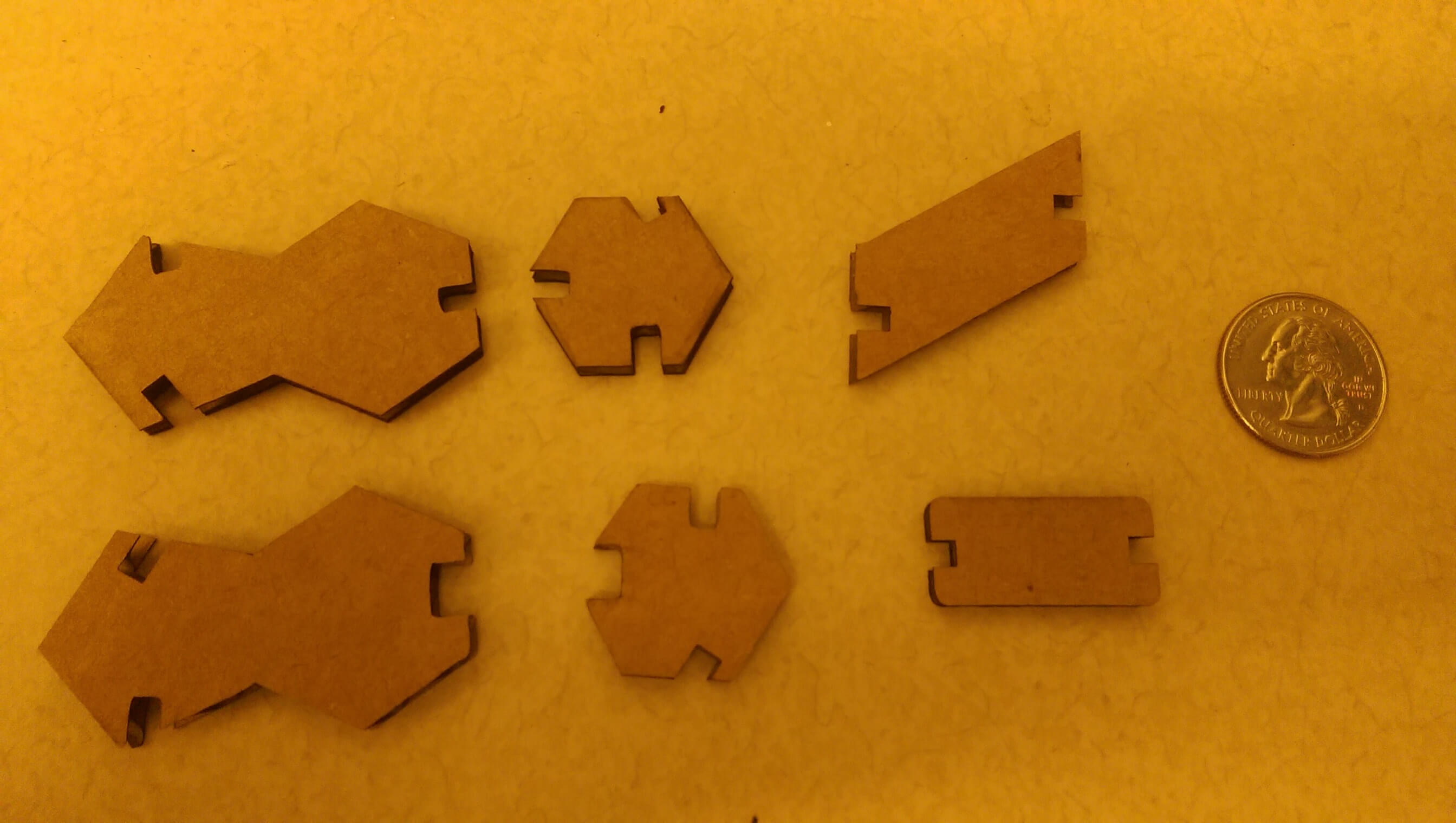
construction kit pieces, CW from top left: adenine, thymine, backbone unit, h-bond, cytosine, guanine

3 assembled DNA base pairs

8 assembled DNA base pairs
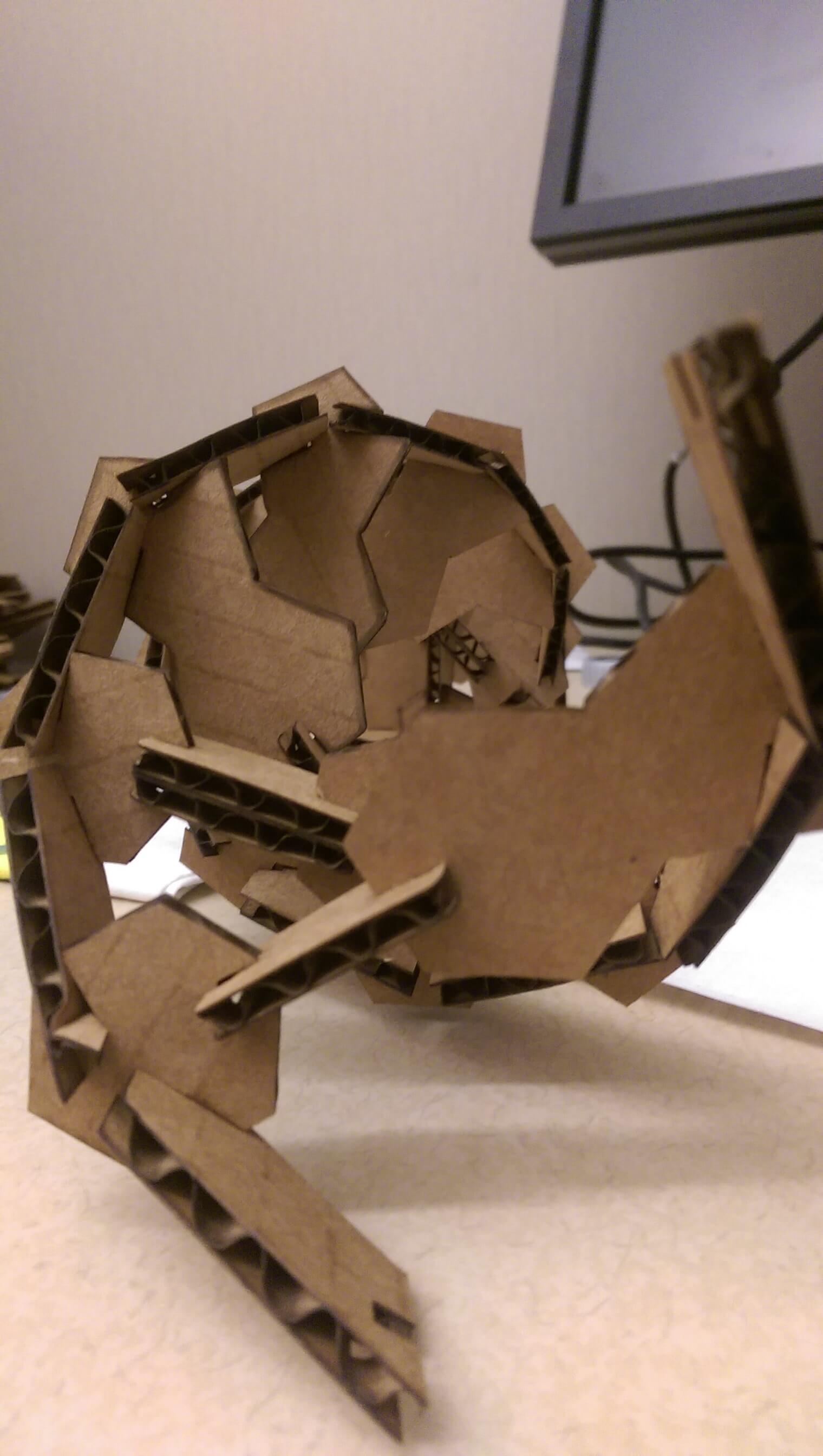
My first iteration worked well enough, but it was not very robust - after about 5 or so base pairs, the structure because unstable and fragile (I could not assemble more than 8 BP in sequence, and even that was unreliable). I decided to do another 'sprial' of my design by tinkering with some design parameters. In this interation, each successful base pair unit needed to be placed under strain to meet the phosphate backbone bonding geometry of its nearest neighbors. This was in part by design - the restoring force of cardboard against this strain contributes to the press fit. However after assembling the kit it because clear that, in this embodiment, it was destabilizing the structure.
I tweaked the design by 1) scaling up the size of the entire kit, 2) realigned the backbone bond vertex to intersect with the hydrogen bond axis and 3) I deepened the cuts for the backbone bonds.

redesigned template
The performance of this kit was *much* improved. Structures were now stable up to ~15 base pairs, and it survived several basic (accidental) fall tests. Moreover it recapitulated some important aspects of the true DNA double helix structure - that is, it admits 10 base pairs per helical turn.

laser cutting the updated press-fit DNA kit

one full turn of the cardboard DNA helix