HTGAA 2021: Bio Production (DIY Gingko)
Guest Lecturer : Patrick Boyle (Ginkgo Bioworks)TA : Pat Pataranutaporn (MIT), Sebastian Kamau (MIT), Eyal Perry (MIT), Alex Mijalis (Harvard)
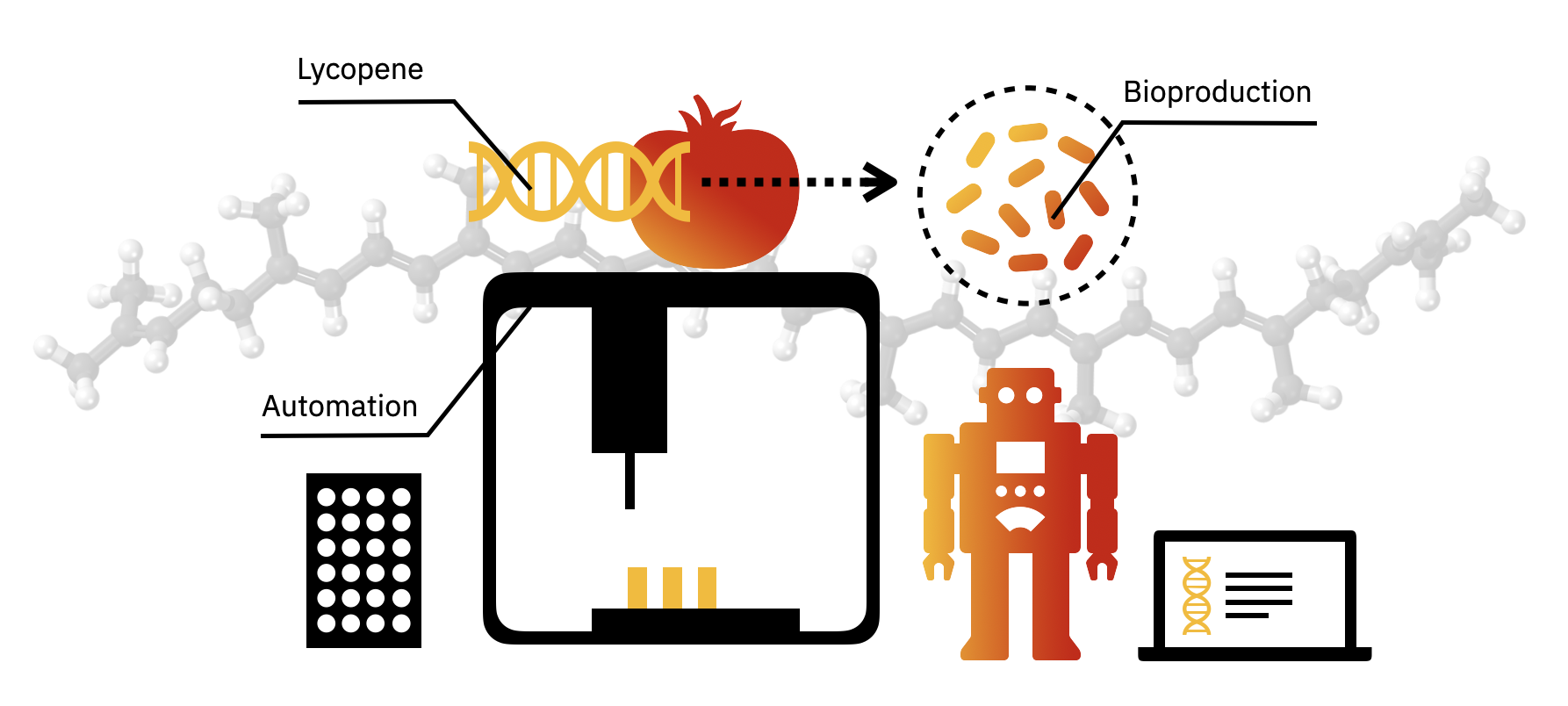
Background
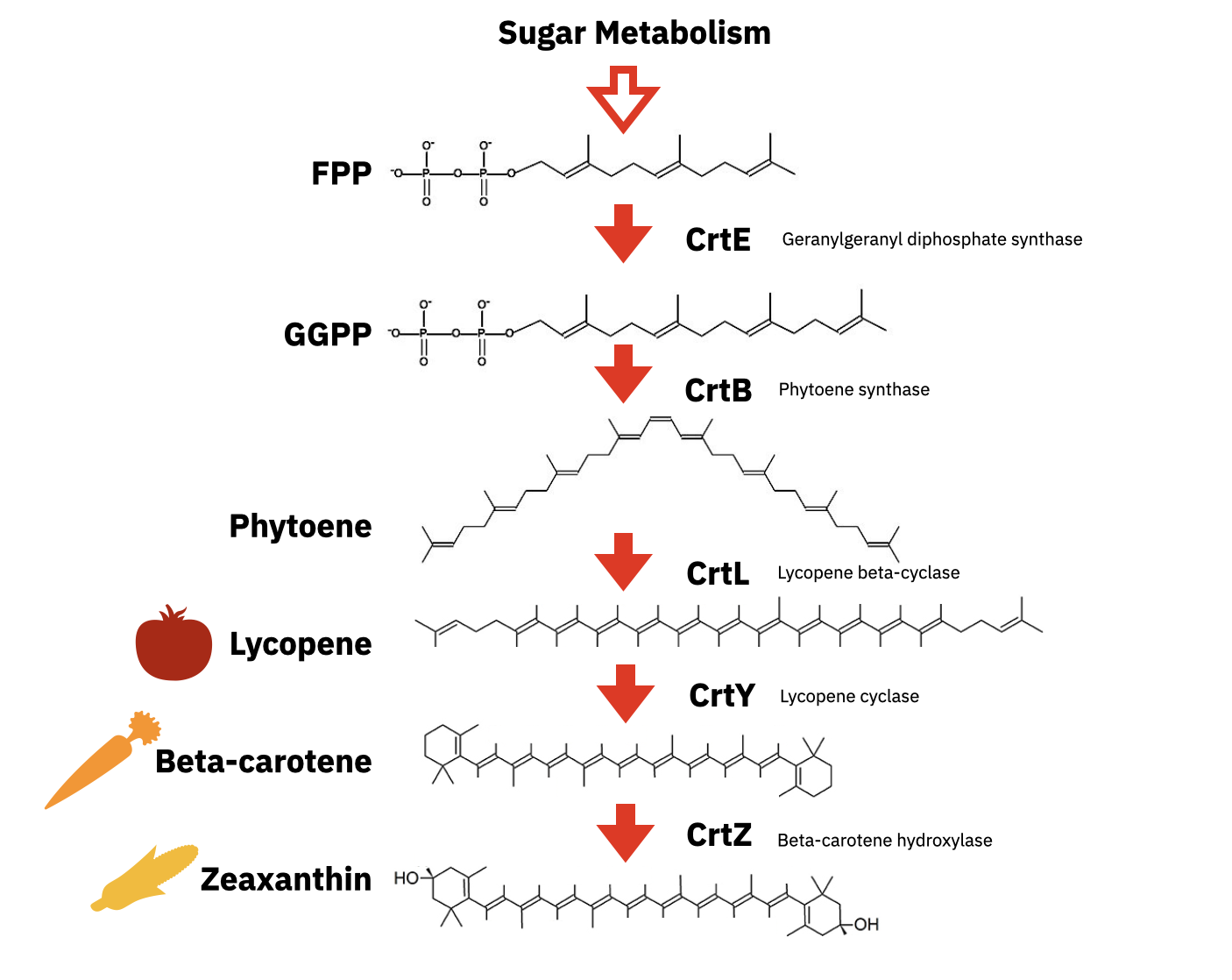
Lycopene is the red pigment that gives tomatoes their color. This pigment is also made by microbes. In fact, transferring a 3-enzyme pathway to E. coli can convert farnesyl diphosphate (FPP) to lycopene. The computational tools and databases presented today can also be used to enhance lycopene production in E. coli or even produce different colors, such as the beta-carotene pigment that makes carrots orange.
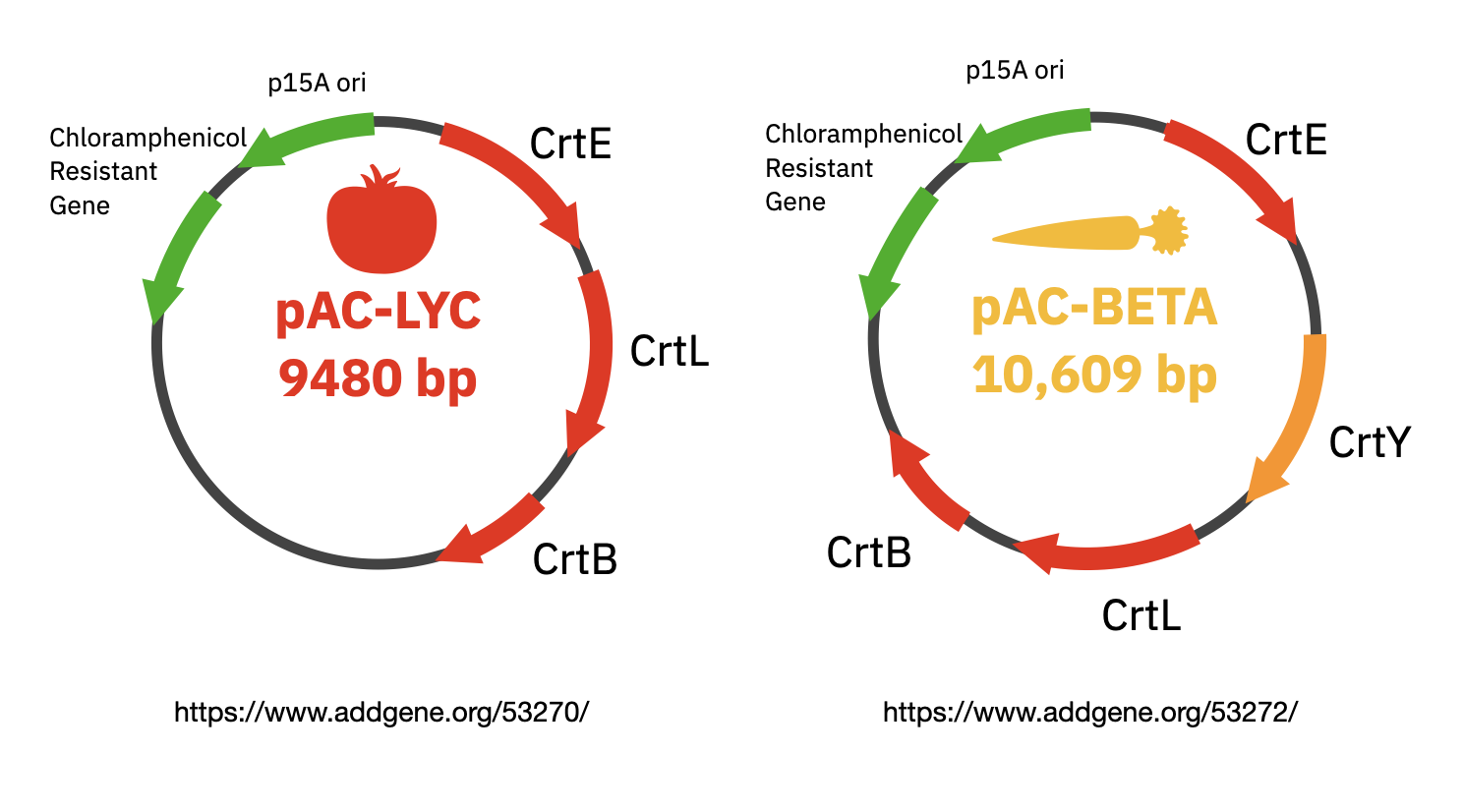
For the laboratory portion of this assignment, you will characterize lycopene production in the E. coli strain from Addgene with the pAC-LYC plasmid. You will also characterize beta-carotene production in E. coli from Addgene with the pAC-BETAipi plasmid. Note, the pAC-LYC plasmid contains three genes from Erwinia herbicola: CrtE, CrtI, and CrtB. The pAC-BETAipi plasmid produces beta-carotene through the addition of the Erwinia herbicola crtY gene. All plasmids include the gene for chloramphenicol resistance.
Assignments
Part 1 : Design and run an experiment to understand how different factors influence bioproduction.
Part 2 : Identify a cool gene (related to your final project) and order it through Twist Bioscience.
Part 3 : Write a short paragraph responding to “If you have the capability of Gingko Bioworks foundry, what would you do and why?”
Recitation Slides
- Plasmid 101 - Sebastian’s Slide
- Homework Overview - Pat’s Slide
Part1 : Tele-Experiment Homework
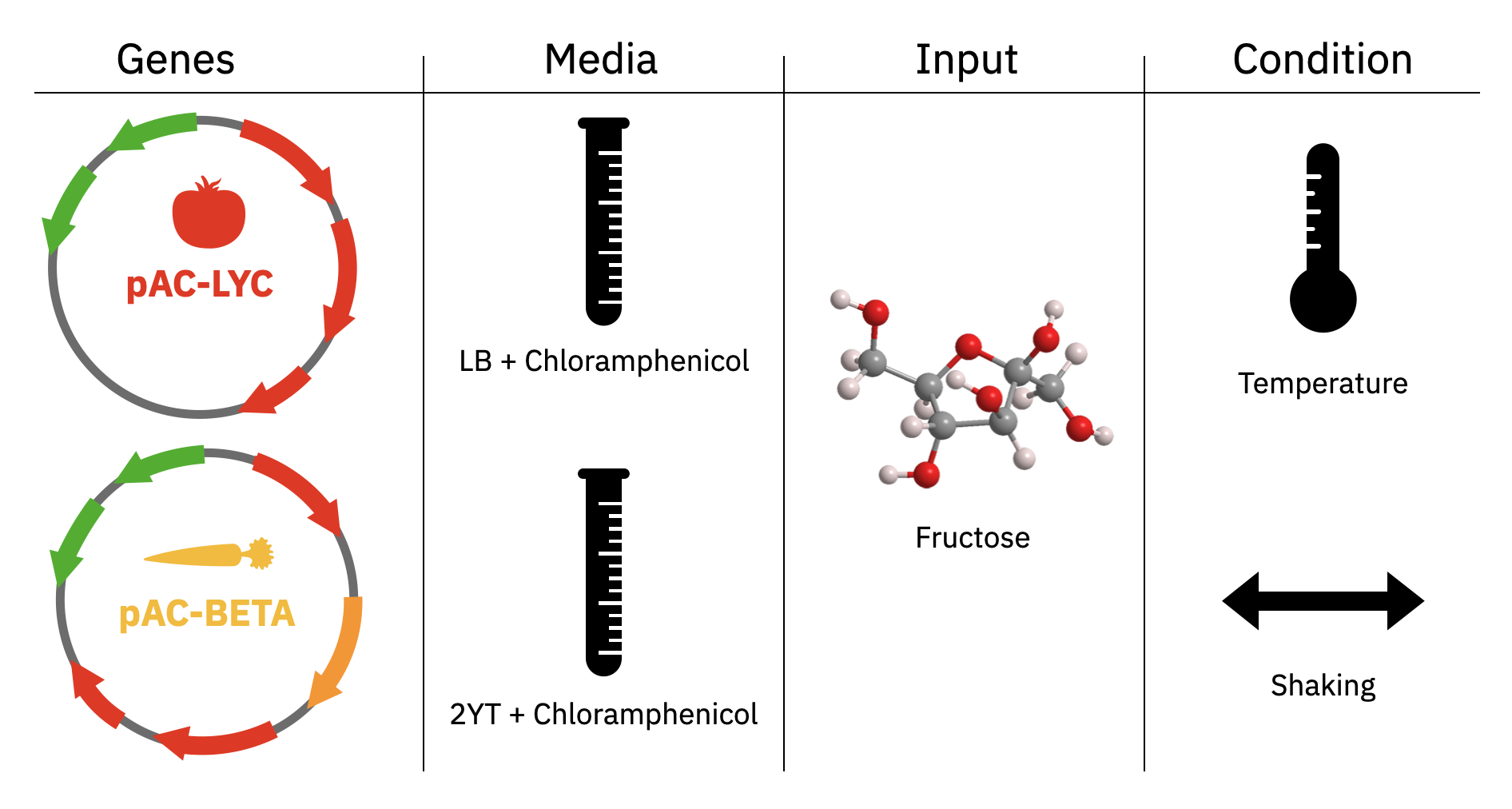
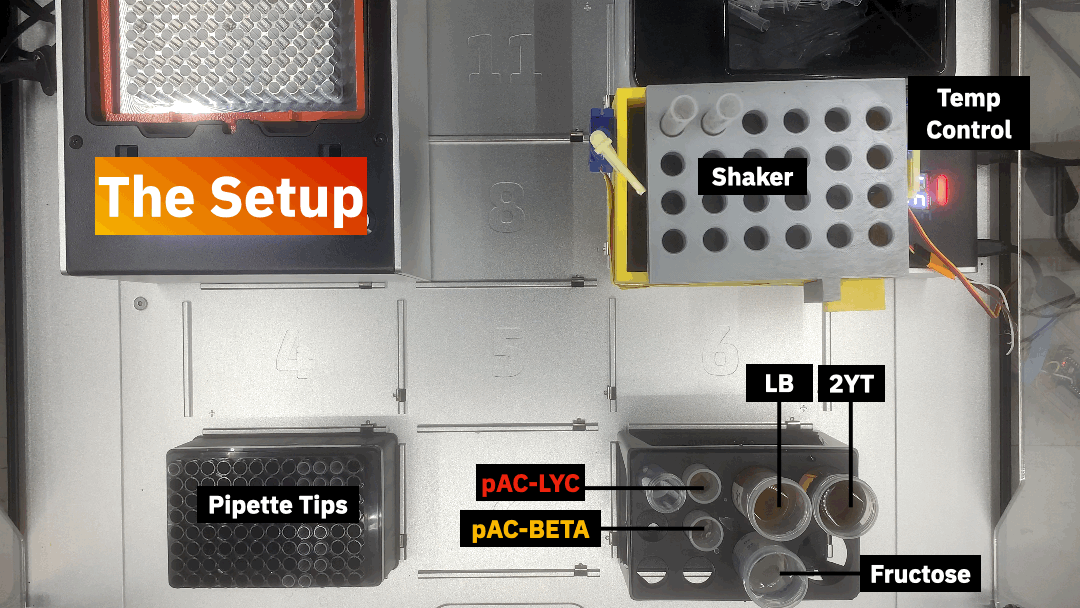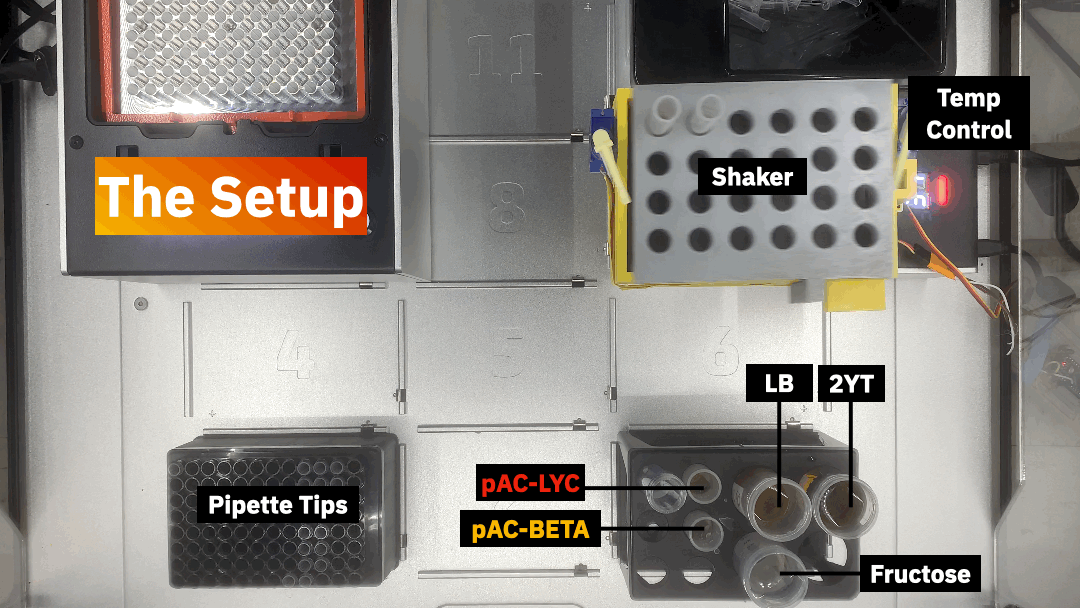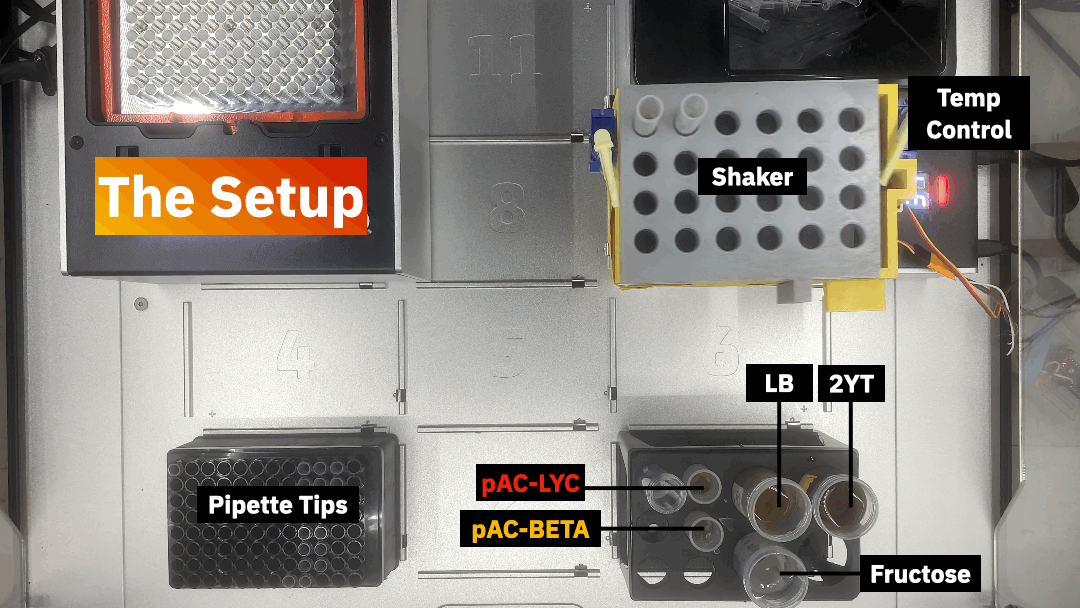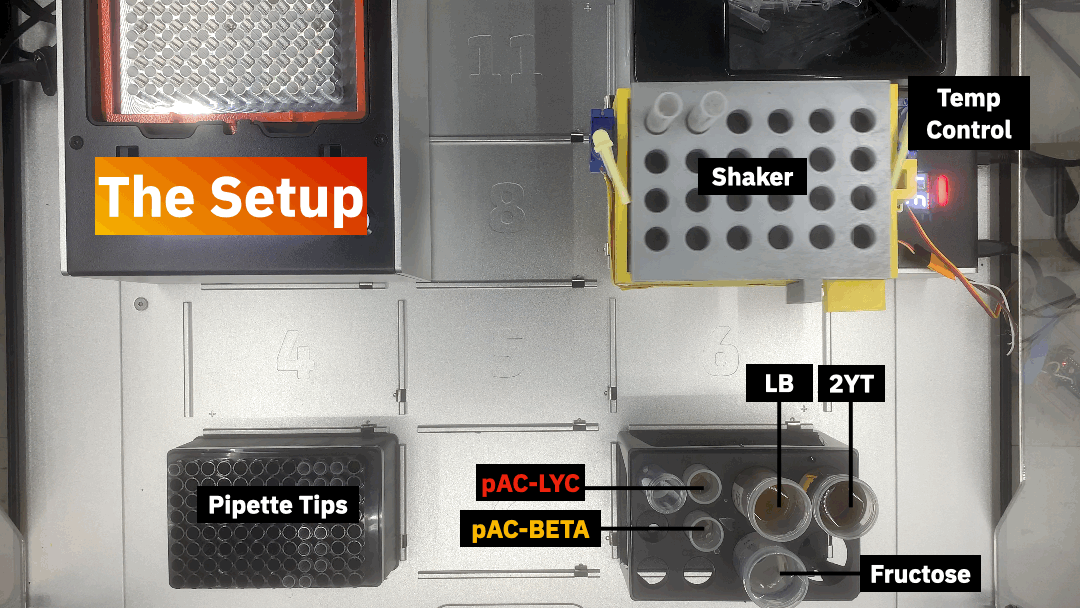
Design and run an experiment to understand how different factors influence bioproduction.
The available factors are genes (pAC-LYC, pAC-BETA), media (LB, 2YT), concentration of precursor (fructose), temperature, and shaking. Please follow the steps below to execute the homework:
- A) Design the experiment
- B) Create the code for the tele-experiment on the robot
- C) Run the simulation to make sure that there is no syntax error
- D) Schedule a time with Pat for running the code on the robot!
Template
The template for experimental design & code for running the experiment on the robot is available here (please make a copy of the file to your own google colab) : https://colab.research.google.com/drive/1pDFV0ADd6JsdiVtcbnKFKFQVVU0NjLVb#scrollTo=sBrB_Z0rVxIg
Scheduling
The student can book the slot to run the experiment on the robot via : https://docs.google.com/document/d/1QwQ3-uxkhSwQnZfEciwykrY10r0up1ikrWt2KNn7btk/edit?usp=sharing
The student must submit the experimental template file to Pat by patpat@mit.edu at least 3 days before the slot, and join the zoom call during the time slot to oversee the robot running the experiment (incase that code contains error/bug) : https://mit.zoom.us/j/93072191099
Part 2: Identify a cool gene (related to your final project)
Twist Bioscience Video Instruction
Part 3: If you have the capability of Gingko Bioworks foundry...
Part 3: If you have the capability of Gingko Bioworks foundry...
Write a short paragraph responding to "If you have the capability of Gingko Bioworks foundry, what would you do and why?" These could be COVID19 related projects or broader synthetic biology projects. Ginkgo Bioworks is taking proposals to leverage the use of their platform to support technical projects; how might you leverage Ginkgo’s technical platform to support your project? Write a description of how you would utilize their platform. What tools and capacities would you use? How would you use them? How would the use of Ginkgo’s platform accelerate or increase the technical capabilities of your project?
Related Readings & References
Blogposts and other resources
- Ginkgo's paper : Development of an engineered probiotic for the treatment of branched chain amino acid related metabolic diseases
- Ginkgo-COVID19
- Synthesize DNA to fight COVID-19
- COVID-19 DNA synthesis project update
- Scaling up whole genome sequencing of COVID-19
- Ginkgo-Moderna-COVID-19
- DNA constructs on GitHub
- News article on the genetic analysis and covid spread of COVID-19
Other Useful Resources
- Gingko’s Paper
- COBRA software for metabolic modeling - not required for assignment
- Lycopene Production Papers for Assignment
- Analysis and expression of the carotenoid biosynthesis genes from Deinococcus wulumuqiensis R12 in engineered Escherichia coli
- Gene expression pattern analysis of a recombinant Escherichia coli strain possessing high growth and lycopene production capability when using fructose as carbon source
- Improvement of Biomass Yield and Recombinant Gene Expression in Escherichia coli by Using Fructose as the Primary Carbon Source
- Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis
- Bitesize Bio Links
- Acetone Precipitation
- E chromi iGEM Cambridge 2009