Week 5 - Measurement and Imaging
Foldscope Assembly
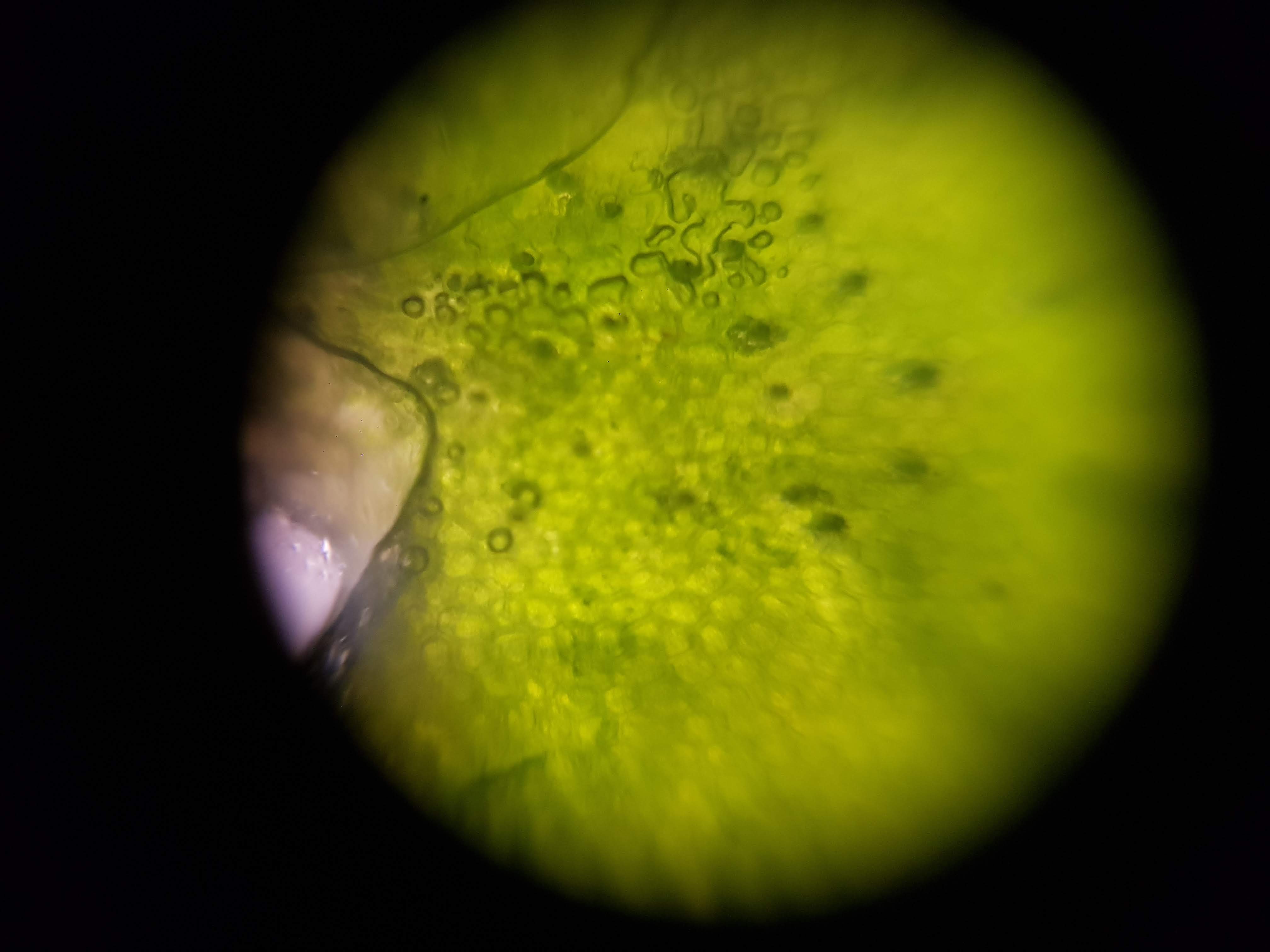
I started this week by building/flofing my Foldscope. The foldscope is an amazing tool that spun out of the CBA. It is a low cost foldable microscope that has 140X magnification and 1.9 microns of resolution!
After assembling the device I decided to test it out with some samples that I got from a cactus friend that was hanging out in the lab. Here are the first images! Its truly amazing how you can manage to distingush different cells and in some cases, even see a little nucleus within them.
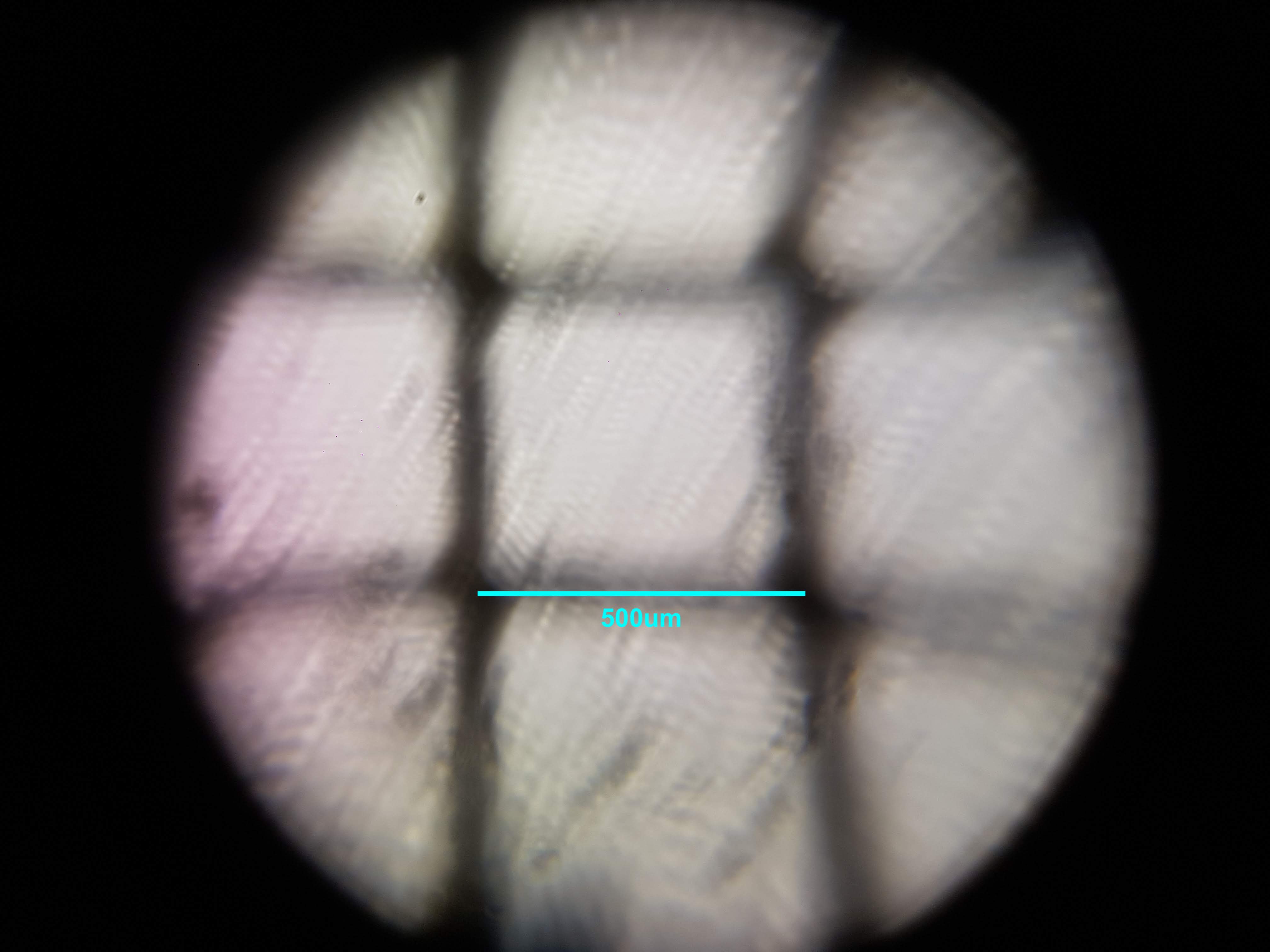
Cactus Sample
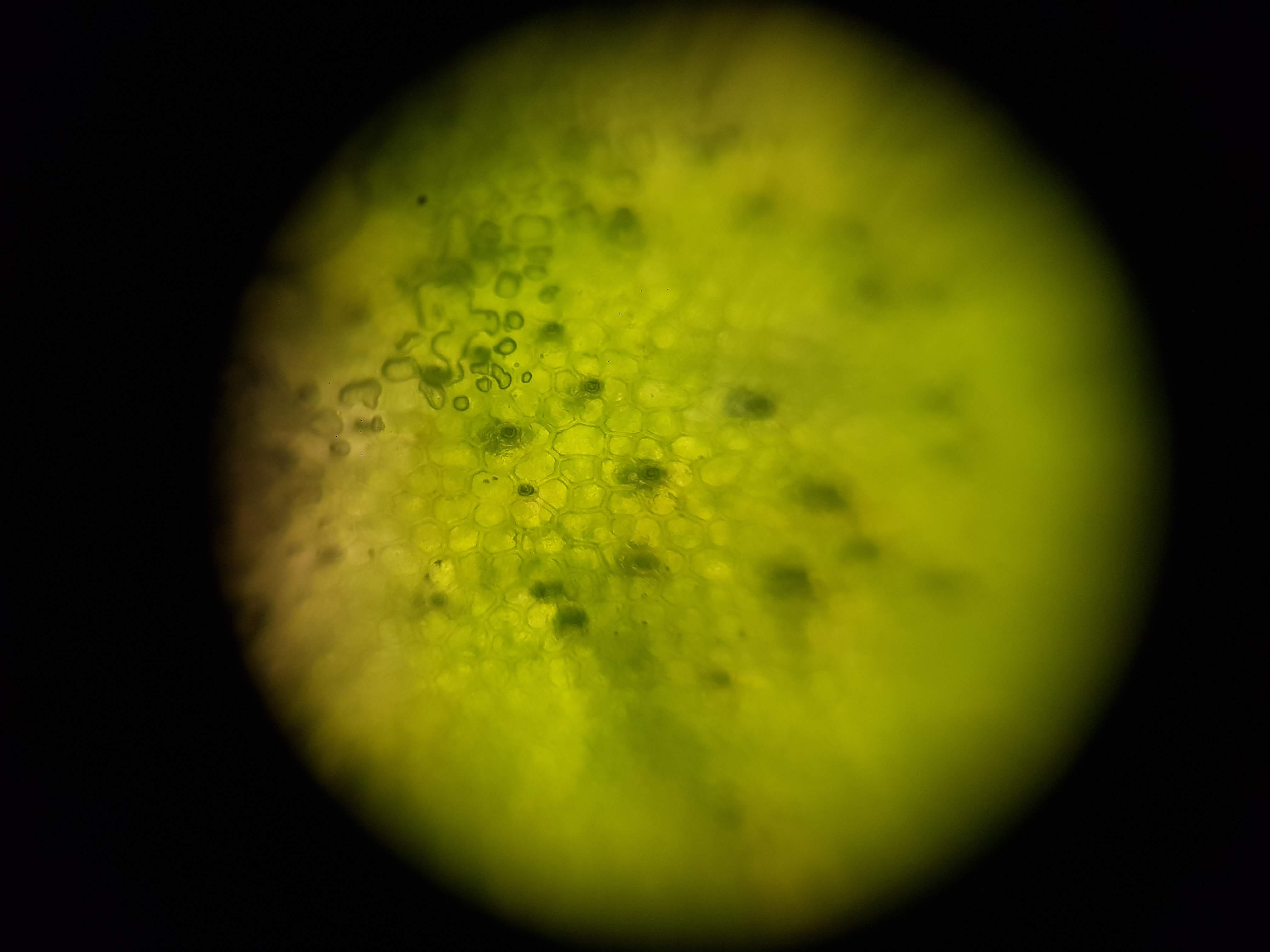
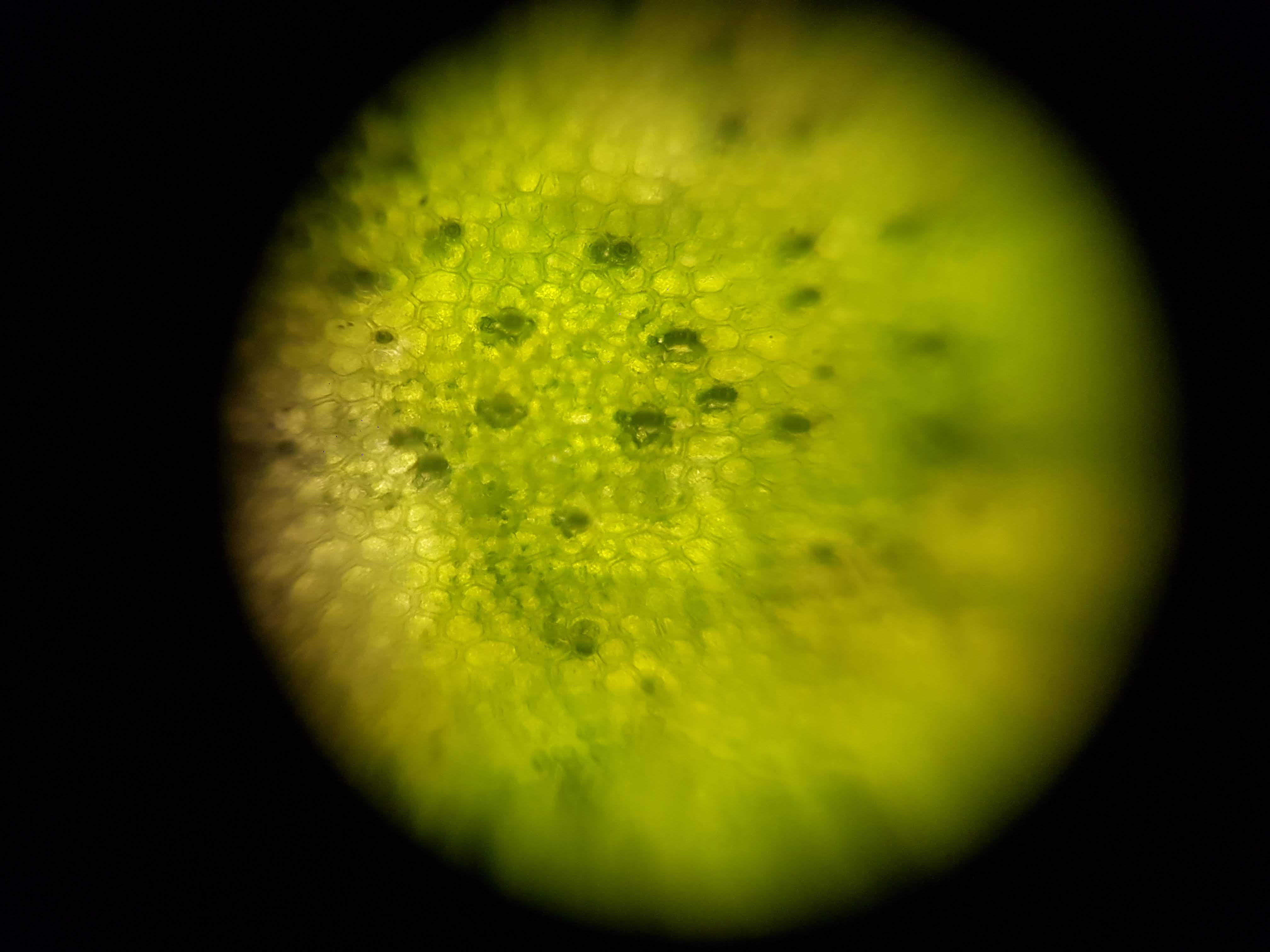
Broadbean Sample
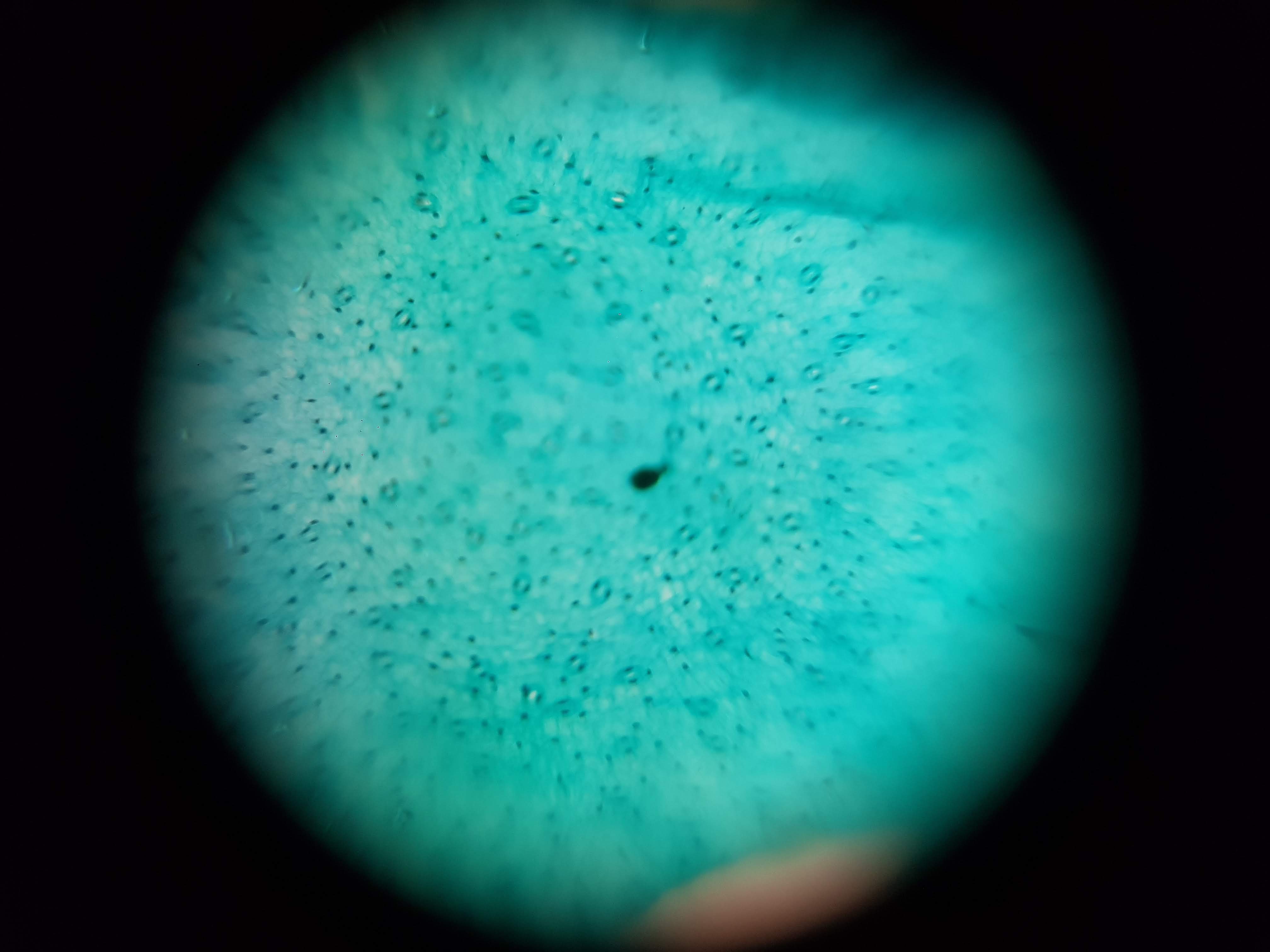
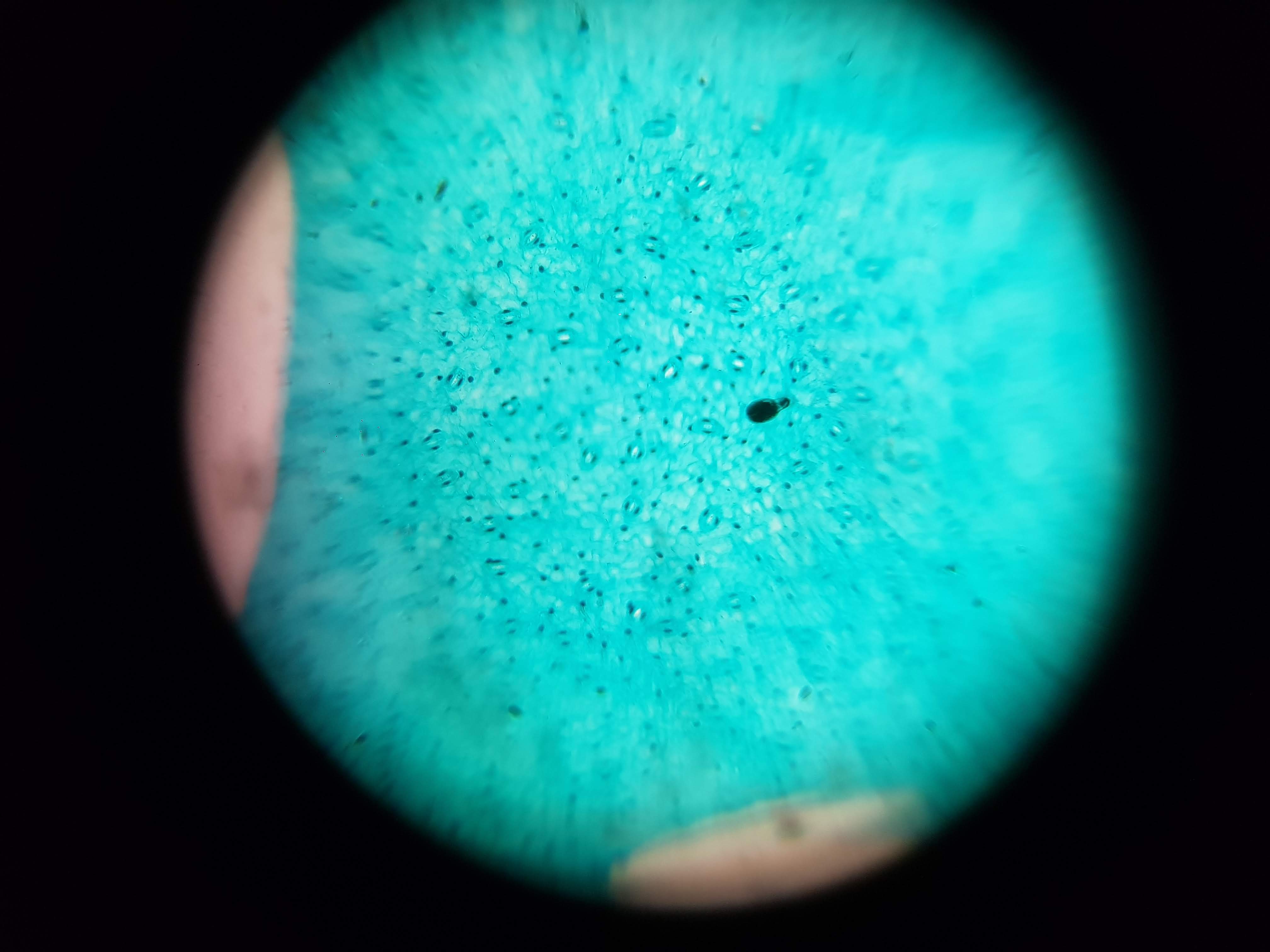
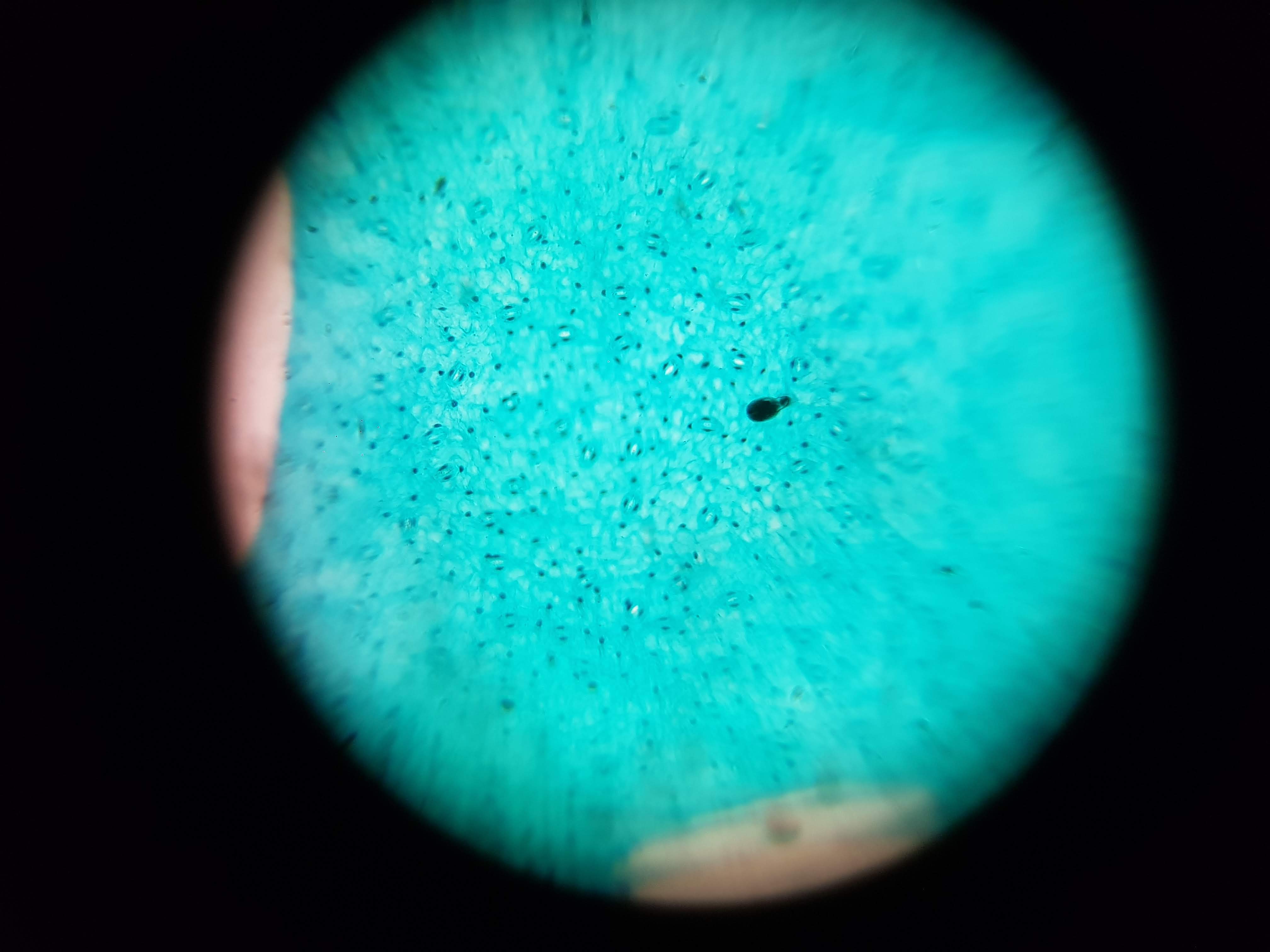
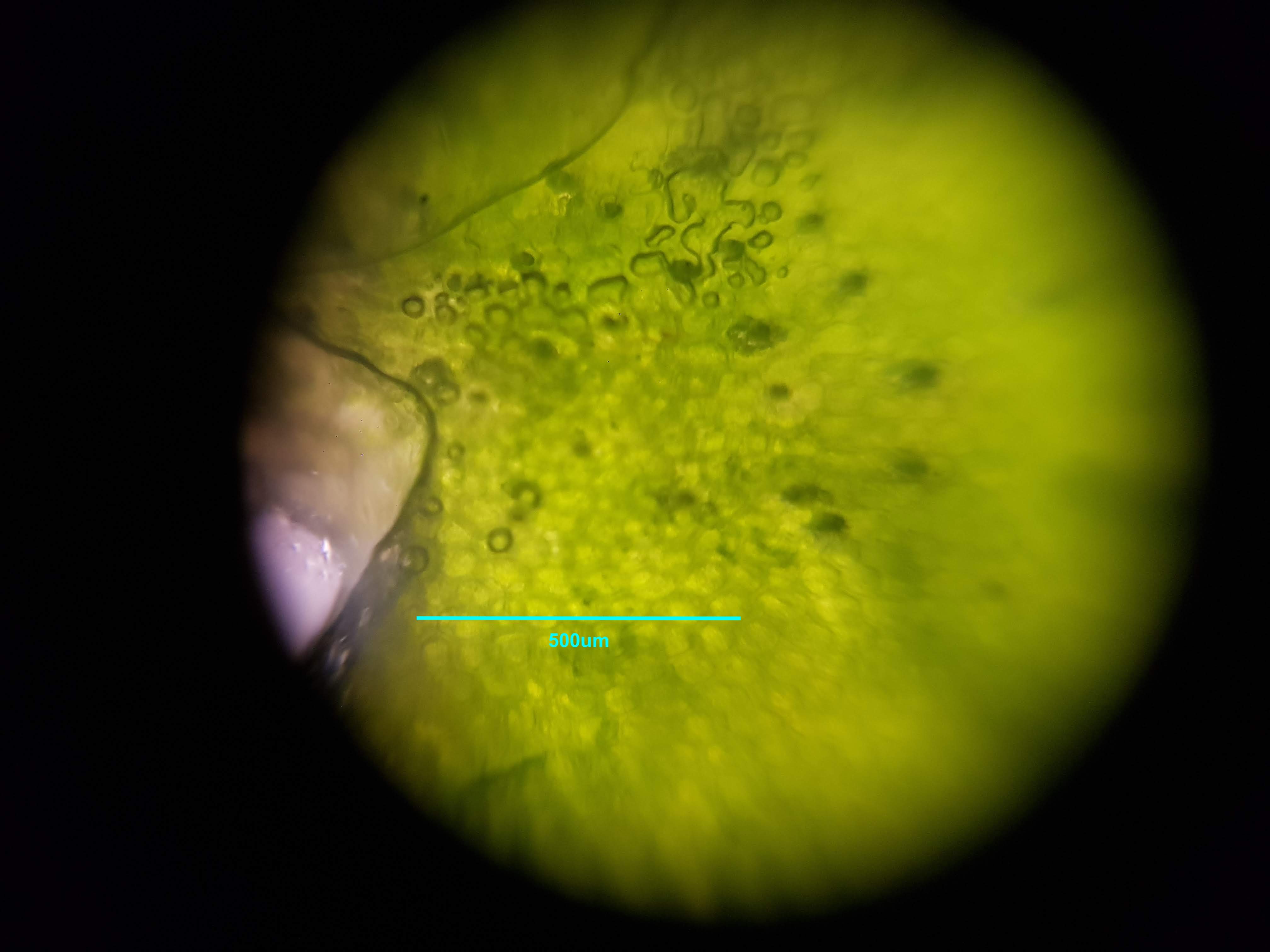
Spirulina Major Sample
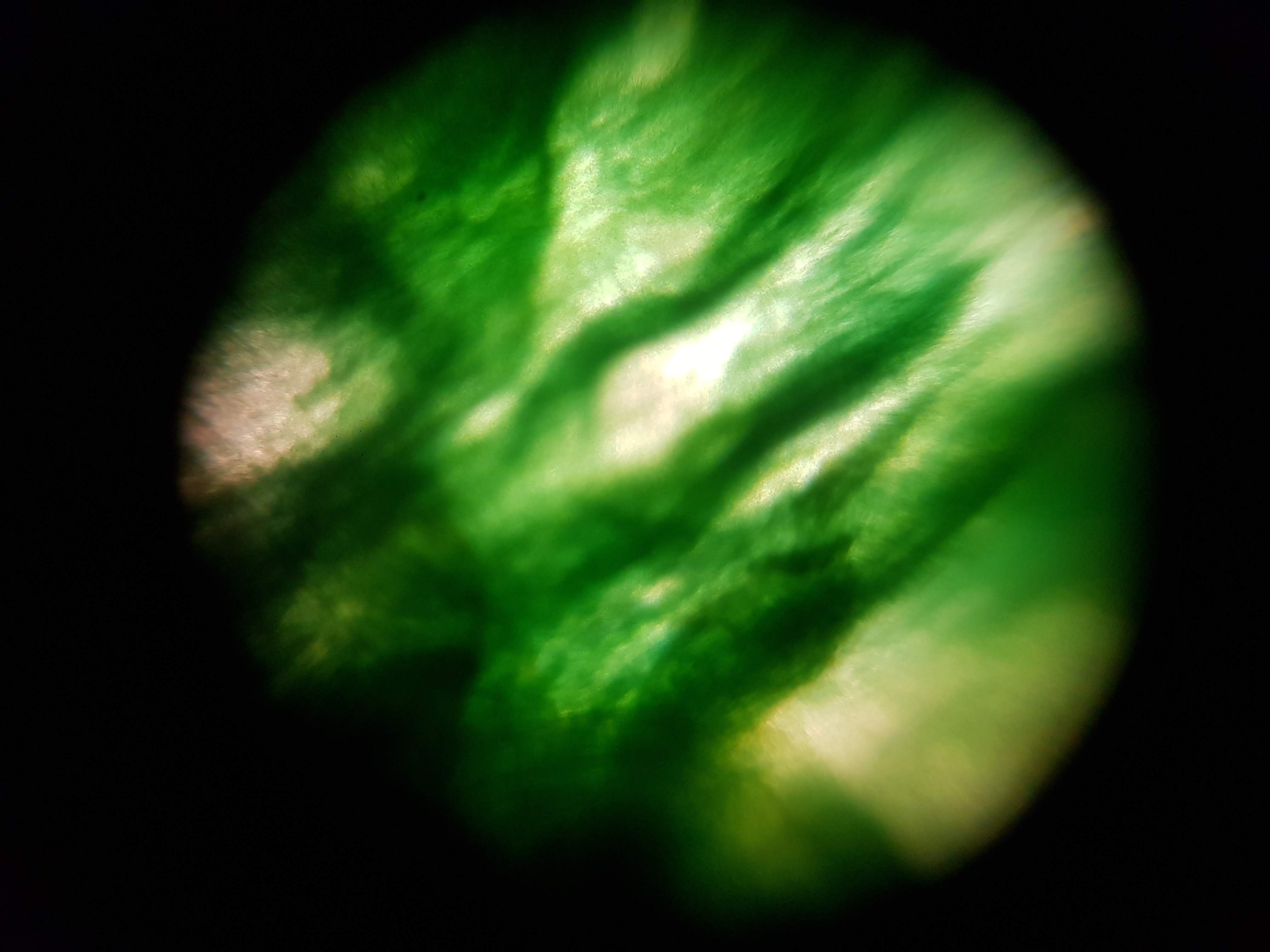
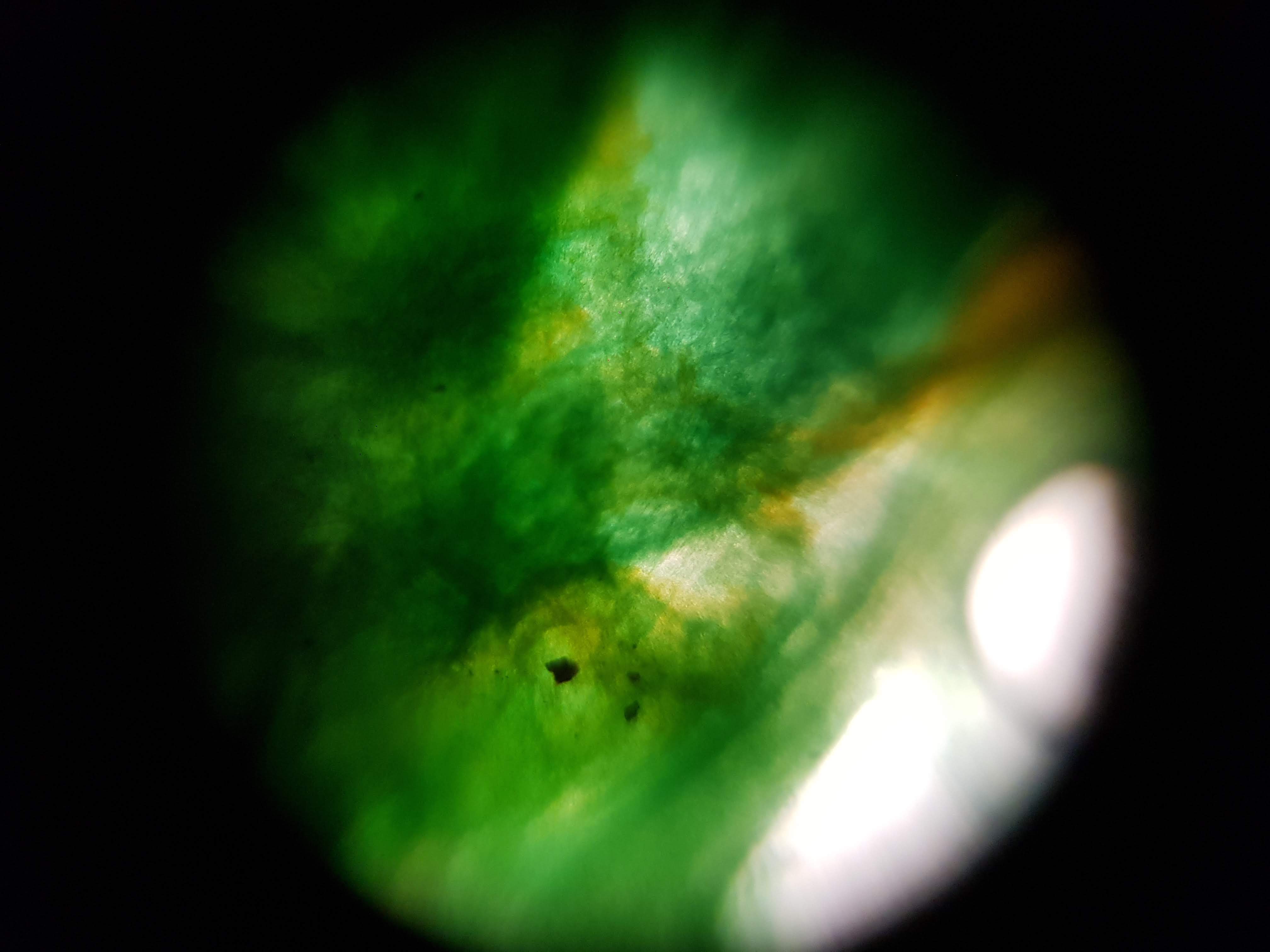
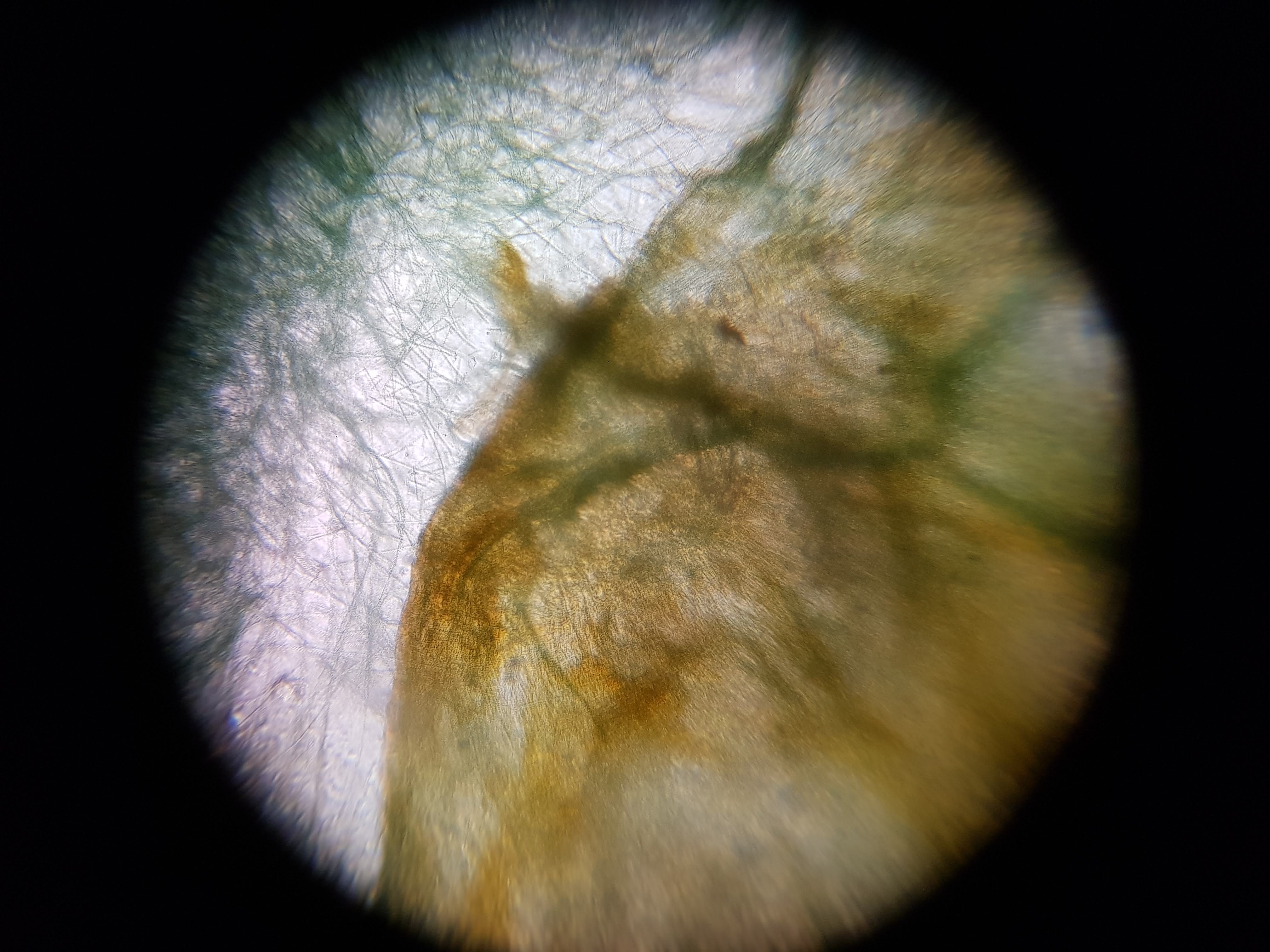
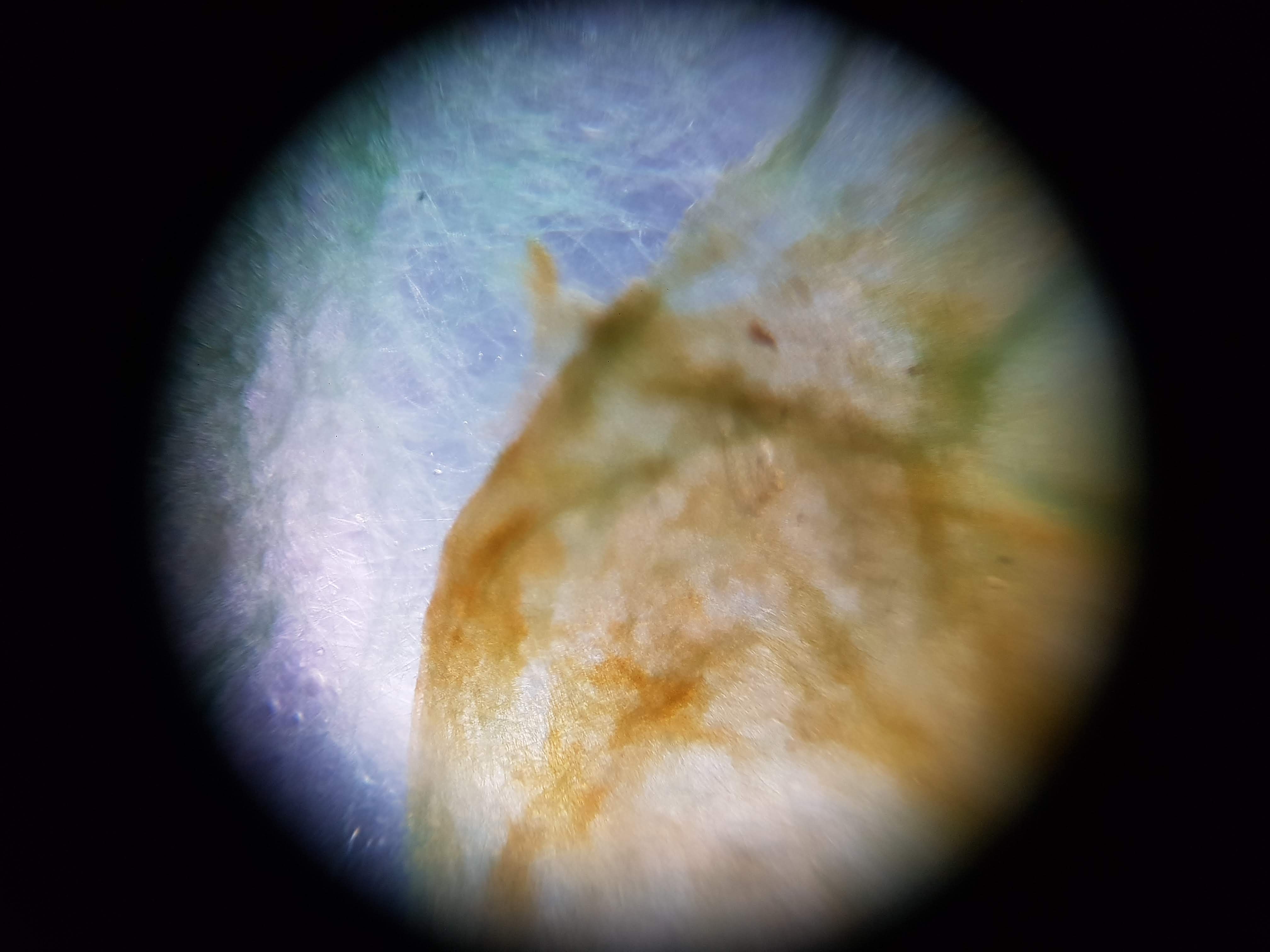
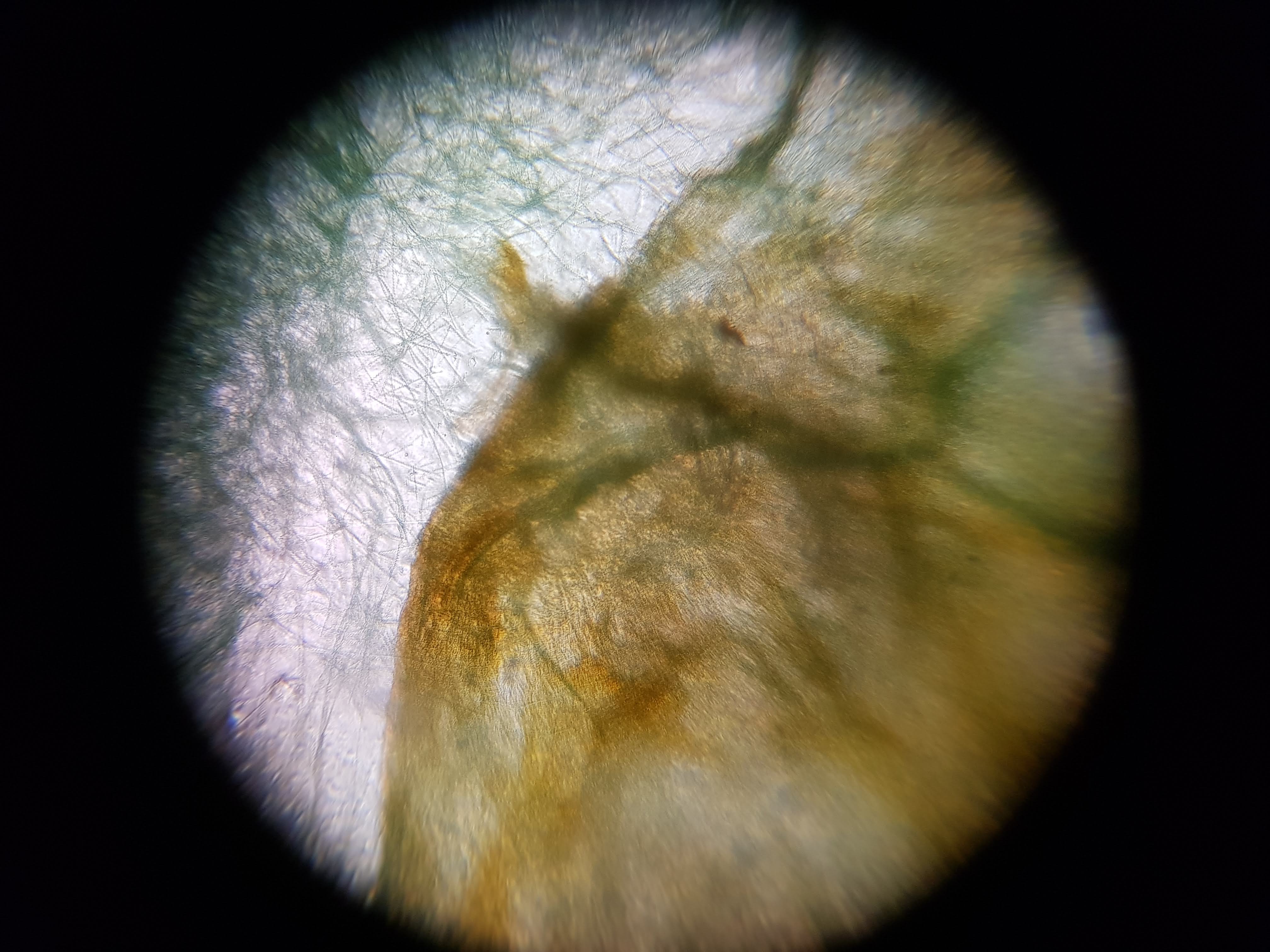
Collection of experimental results:
- I was interested in seeing some comparisons of how different plants and algae looked like. The first sample was a cactus sample that I took from my home. I wanted to see all of the water that it can store internally and to find out if there is anything interesting about its internal structure that helps it retain those liquids. Interestingly I realized that the cells on the cactus sample seemed to be much larger than the ones on the boradbean sample (second row of images). I would attribute this to the extra water that the cactus can hold but it could also be that the cactus sample was a fresh sample as oposed to the broadbean sample which came from the foldscope kit.
- What did you observe? I got to see different membrane sizes and I think I was also able to spot some nucleuses in the sample. I was also amazed at how interesting the interface between the green and brown elements from the spirulina is. The green elements seem to be pulled towards the brown structure and they seem to be hugging it. I wonder what thats about?
- Magnification: I used a nice online magnification calculator. I used the tech shit from my phone to know the display pixels as well as the sensor
pixels. I gave the following input to the calculator.
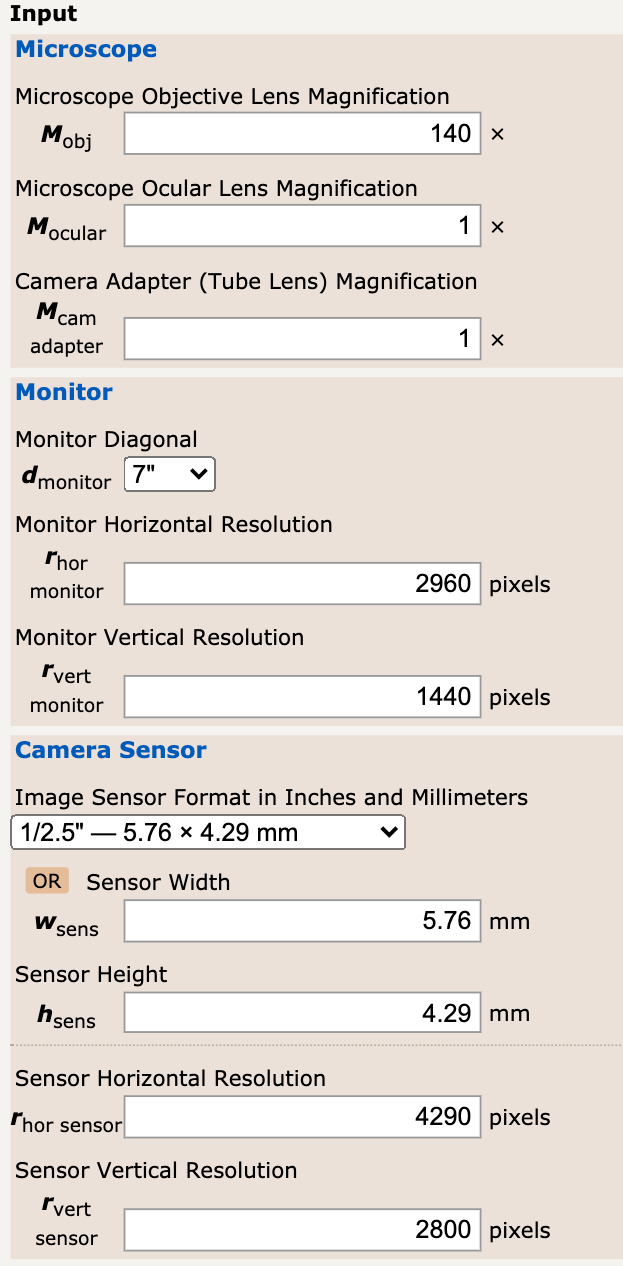
I ended up getting a final magnification of M = 5,632.17X It seems amazing that we are able to get this type of magnification with such common tools! In terms of smapling frequency, it should depend on the specifications of the device that you are using to capture the images. Phones should be above 100Hz.
And since I want to be a good scientist, here is a picture of how I calculated my scale bar!
- Foldscope is very unique in the sense that the optical system is composed of only one spherical ball lens as oposed to more traditional and common devices that use a complex system composed of multiple lenses. This allows foldscope to have a magnification of 140X within slips of paper. Another amazing characteristic of foldscope is that the microscope is able to useexternal sources of light, commonly, systems have an aligned source of light within them.
- Resolution varies depending on the field of view that you have and can vary depending on your experimental setup. Foldscope, within given scenarios is able to achieve a 2um resolution.
Design of smFISH / Spatial Sequencing Assay
I will atempt to design my smFISH assay guided by the Guide for RNAScope. The excersise that I will be looking to see Heterogeneous target expression in a particular cell type. To do so, I will be looking into the expression of AFAP1-AS1, which can be helpful for diagnosing or detecting human lung cancer. The assay would involve one color fluorescence, if the gene is expressed it could be likely that the patient has cancerous cells. I was able to find my gene and sequence properly!
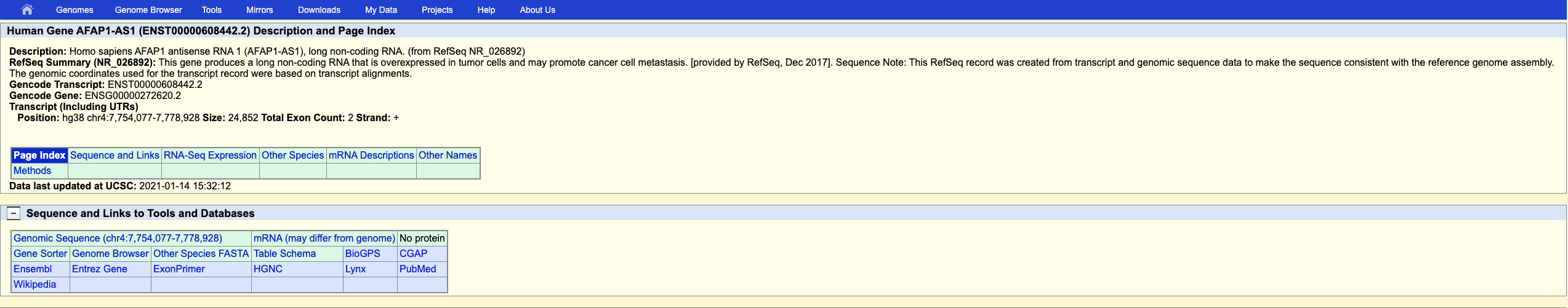

Next, I will start with manually testing 5 different 25 base pair probes to understand their melting temperature and if they have a hairpin.
CTCGTTTTCTTTTGCTGCTGCCACG
Primer Tm:
60.01103488834258
Hairpin details:
ThermoResult(structure_found=True, tm=36.50, dg=30.81, dh=-19100.00, ds=-61.68)
GGCCTCCCCAGCCATGTGCAACTGC
Primer Tm:
66.06598733342202
Hairpin details:
ThermoResult(structure_found=True, tm=46.68, dg=-798.71, dh=-26400.00, ds=-82.54)
CAATGGAGTGACGGCATCCAACTCA
Primer Tm:
59.27299103448951
Hairpin details:
ThermoResult(structure_found=True, tm=41.14, dg=-370.33, dh=-28100.00, ds=-89.41)
TGACTCACATGGGAAGAGGACCCCA
Primer Tm:
60.58207613669754
Hairpin details:
ThermoResult(structure_found=True, tm=51.35, dg=-1335.93, dh=-30200.00, ds=-93.06)
CTTCATTCATTTCAGTTTCCCCAGC
Primer Tm:
54.931318607493836
Hairpin details:
ThermoResult(structure_found=False, tm=0.00, dg=0.00, dh=0.00, ds=0.00)
I then ran a complete analysis and chose the following probes:
Starting base: 513 Sequence: TGTCAGGAACACAGGCCCGGCCGAT Tm: 65.80308921252146 Hairpin Tm: 37.952710437431165
Starting base: 1120 Sequence: ATGCTGATTCTGACGAAGGCCAGGA Tm: 60.00235377725454 Hairpin Tm: 29.39903093017341
Starting base: 4975 Sequence: CATGGCGATTGAGACTCCGTGGGAC Tm: 61.33947044115877 Hairpin Tm: 28.085286313193933
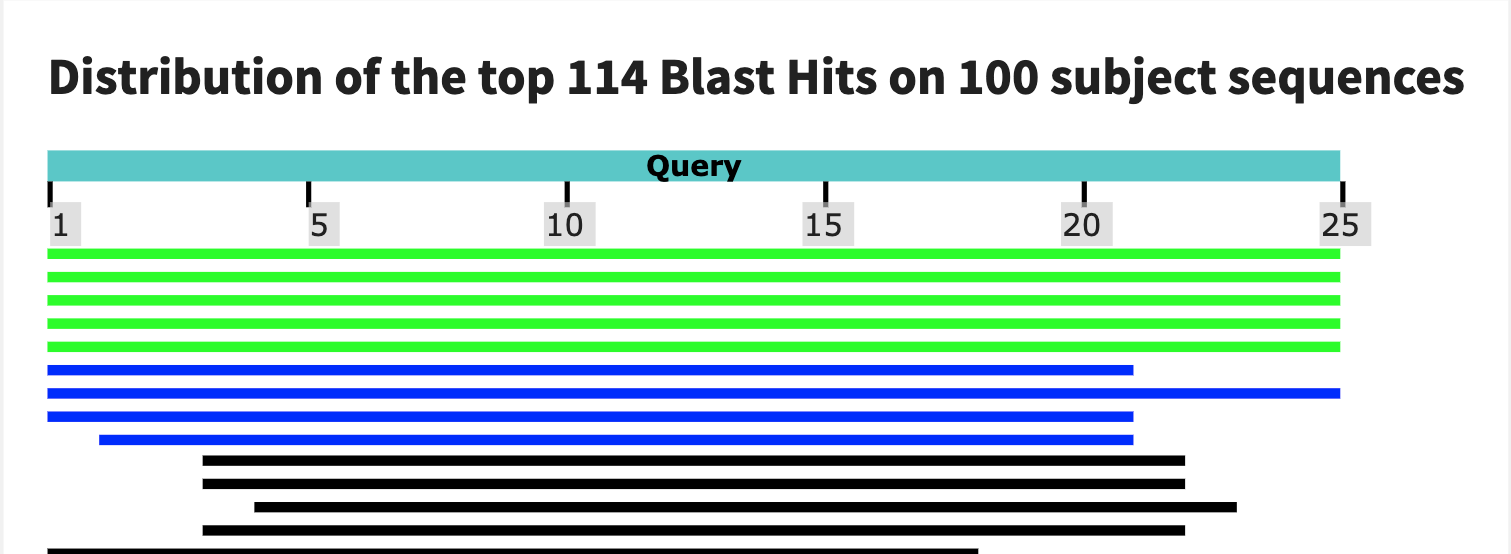
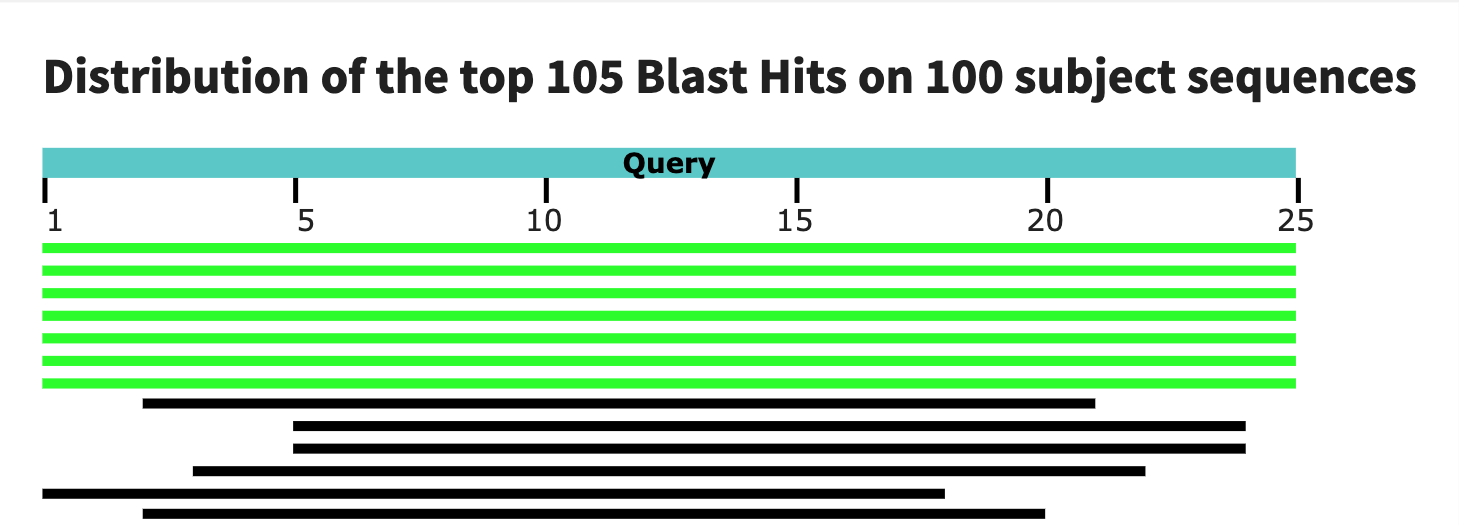
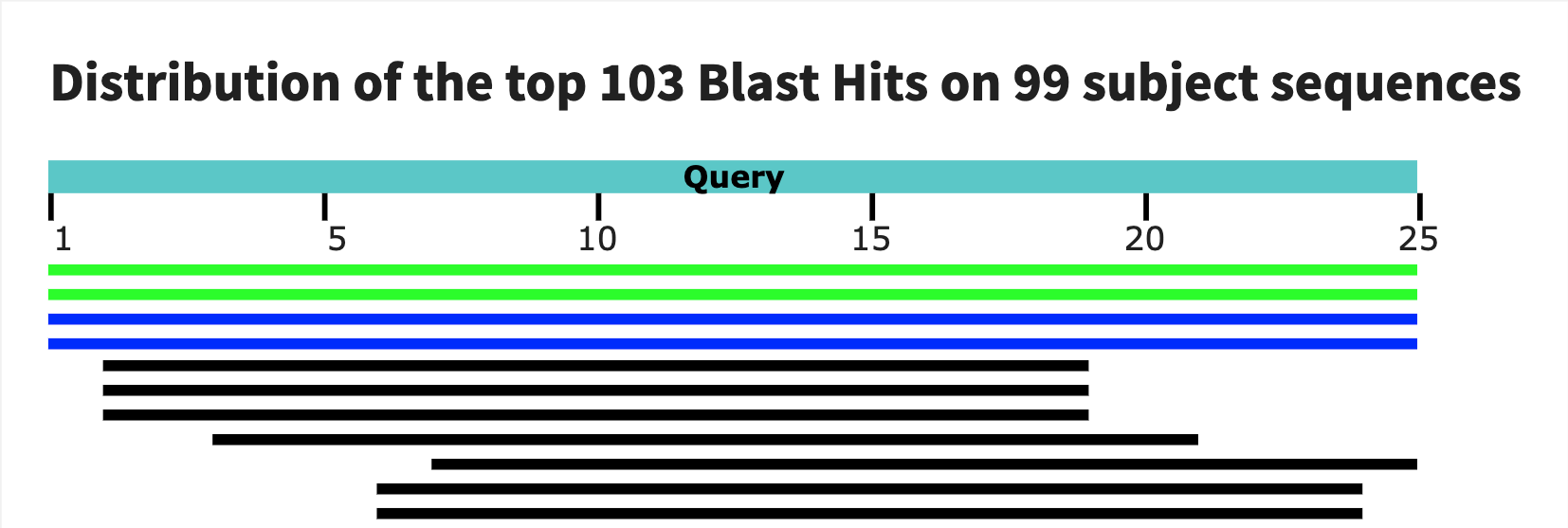
Finally, I used BLAST to test the specificity of my probes. Intresetingly, the first probe has a large match with gorillas! The second one seems to be much more reasonable. The tird blast is not as compelling as the second so I would end up choosing the second as my assay.