Week 6 - Circuits, Sensors & Cell-Free Systems
Part A - In Silico Homework
Picking a Fucntion
As part of this week's assignment we will be designing our own useful synthetic minimal cell. I will try to start aligning this assignment with my final project ideas so that I can start to test out
some of the concepts that I'm thinking through for my final project. For my final project I would like to see if I can build bio based sensors for understanding light or/and sound within the context of public spaces in the city.
There is interesting work around understanding sound-responsive gene growth [1][2][3][4].
1A What would your synthetic cell do? What is the input and what is the output. I would like my synthetic cell to be used as a 'bio microphone'. The idea would be to add sound-responsive genes so that expression can be activated whith exposure of the cells to certain frequencies.
Different sounds can change gene expression and growth rate of my populations. Input -> Sound / Output -> Color. The cchanges of growth and color could later be monitored by a light sensor. This would allow for non invasive sound monitoring within cities.
1B Could this function be realized by cell free Tx/Tl alone, without encapsulation? No, because our ourput depends on cells. If reaction done without the cells, we would have a hard time quantifying and reversing our outputs.
1C Could this function be realized by genetically modified natural cell? I believe this could be done through a natural cell. Most of the genes that have been found to respond to sound have been found in plants and plant cells tend to be harder to work with so
using a synthetic cell seems to be a good alternative.
1D Describe the desired outcome of your synthetic cell operation. In presence of sound stimulation my cell will produce certain color pigments. I suppose the system could also be implemented within live cells so that we can use growth as measuring parameter.
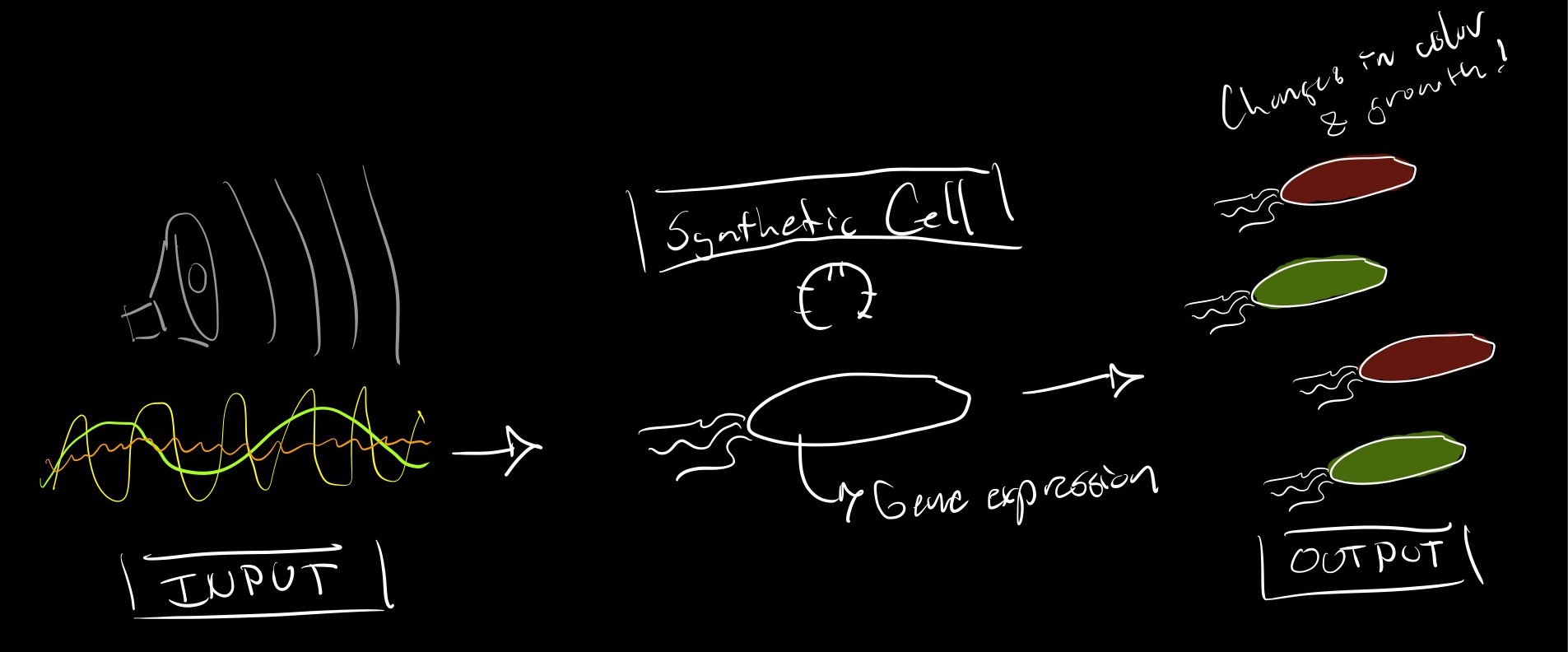
Designing components that would need to be a part of the synthetic cell.
2A What would be the membrane made of? The most common membranes used use phospholipids + cholesterol. Since my membrane will not need to have any specialized communication attributes, using a sympole lipid one should be ok.
In addition, this type of membrane is robust to the frequencies that I'm interested in.
2B What would you encapsulate inside? Enzymes, small molecules. I would encapculate a basic cell free Tx/Tl system guided by sound sensitive promoters and a chimeric gene of intrest. When activated by sound, the promoter would express the CAG sequence from mUAV to express color.
I guess that I would also have to encapsulate the bases needed for my proteins of intrest to be expressed.
2C Which organism your tx/tl system will come from? I would have my tl/tx system come from EColi and from corals as we did two weeks ago. Most of the effects of sound that have been published are observations in plants but I did manage to find examples of the within bacterial cells.
Since bacterial cells are easier to work with I would choose a bacterial tx/tl system.
2D How will your synthetic cell communicate with the environment? The system is quite simple and wouldnt need to interact with its environment to echange inputs or outputs but it would be important for all the normal nutrients and components to be able to go thorugh the membrane so that gene epxression is carried out properly.
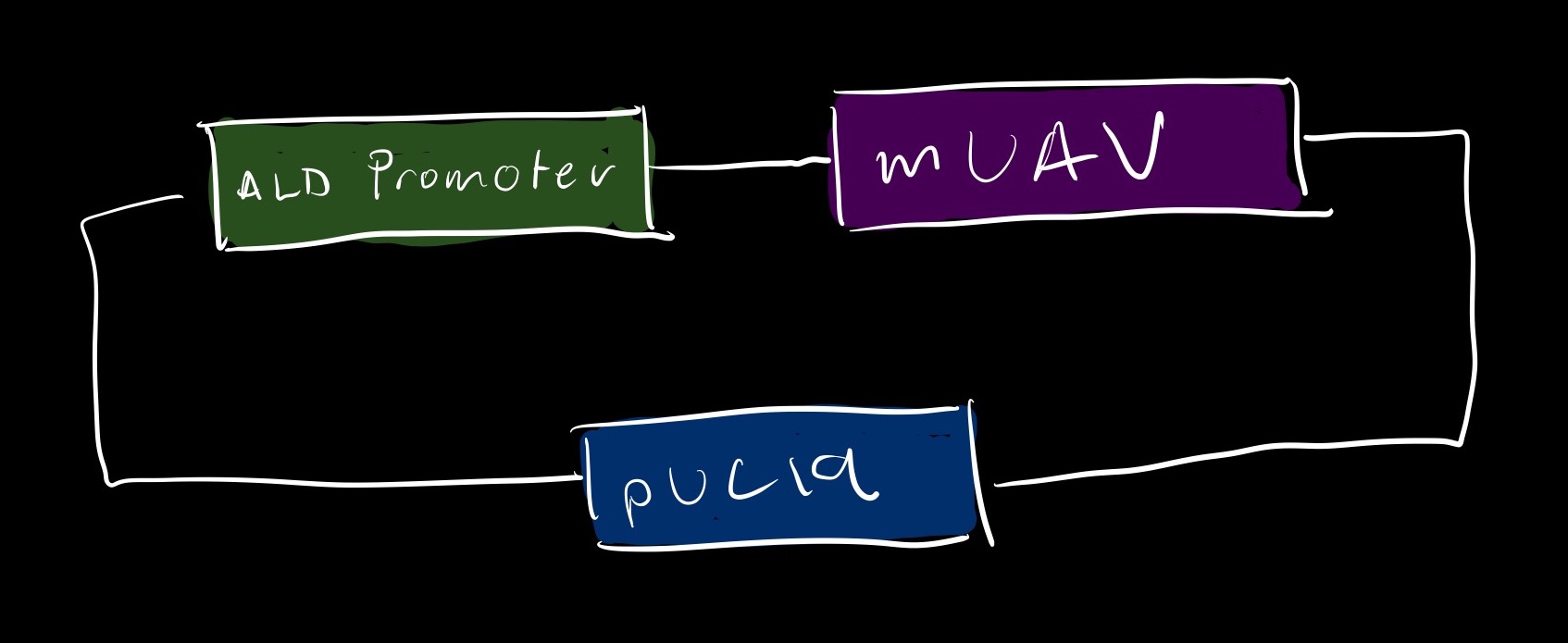
Experimental Details
Enzymes: Bacterial cell free tx/tl
Genes: rbcS & Ald mRNA -> Expression upregulated between 125 - 250Hz and downregulated at 50Hz [1]. CAG sequence from mUAV for coloring [4].
Biological Cells: E Coli transformed with mUAV gene and ALD sound sensitive promoter.
3B How will you measure the function of your system? The function of my system would be measured by using the naked eye but more intrestingly having a color or light sensor that can translate expression into an electronic signal that can then be processed would be the best way.
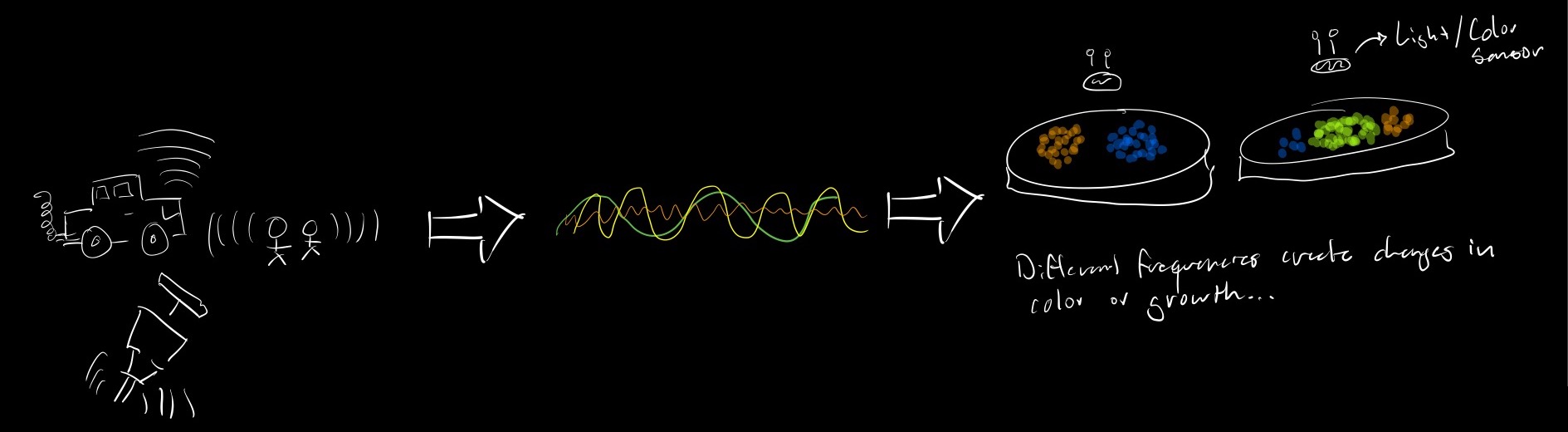
Part B - Experimental Cell-free transcription
This week I will be designing several experiments with the minipcr biobits! Biobits are an MIT born educational tool that is transfroming the way students all around the world engage with synthetic biology. To start of the process I watched this great live demo by Ally to understand what we are to do with this week's experiments. We will be designing a transcription that produces a fluorescent RNA called Broccoli.
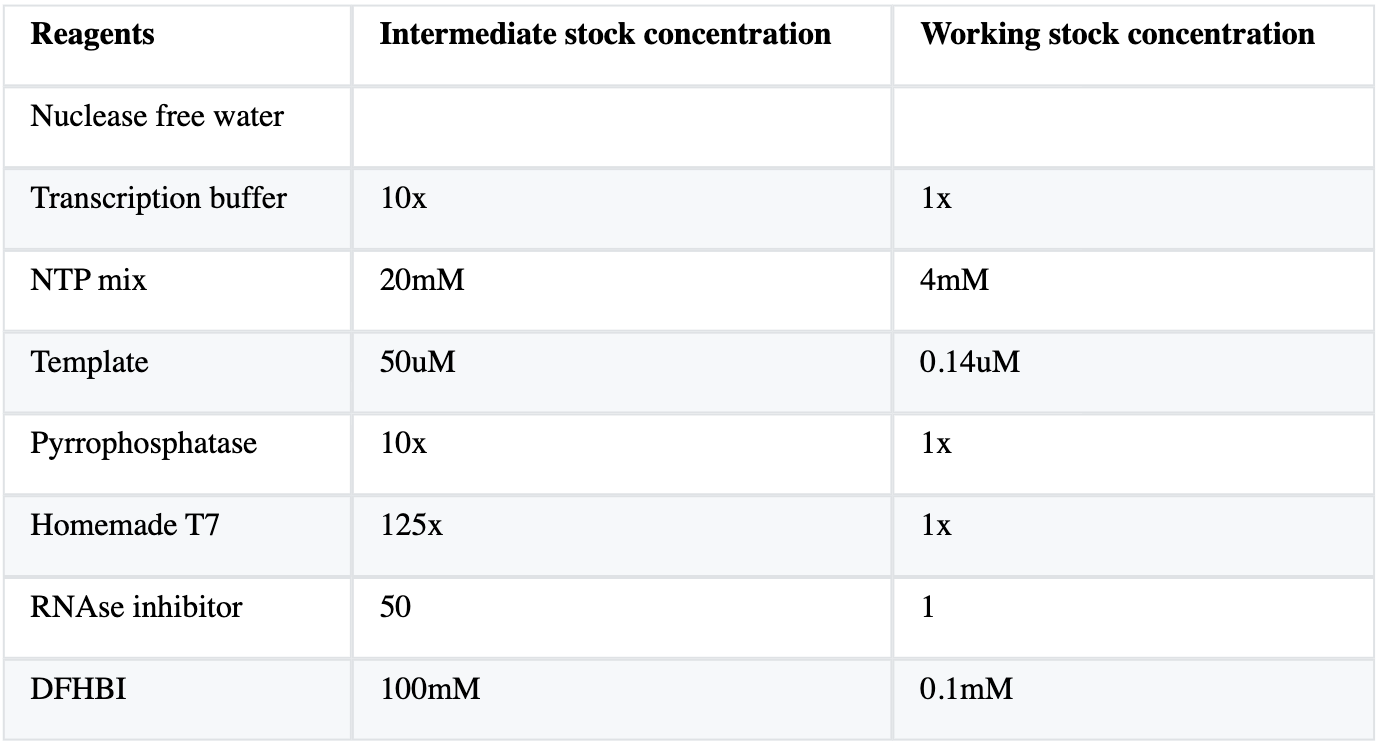
We will start off with the basic cell-free transcription reaction, detailed in the image below:
And then carry out the basic translation reaction outlined below:
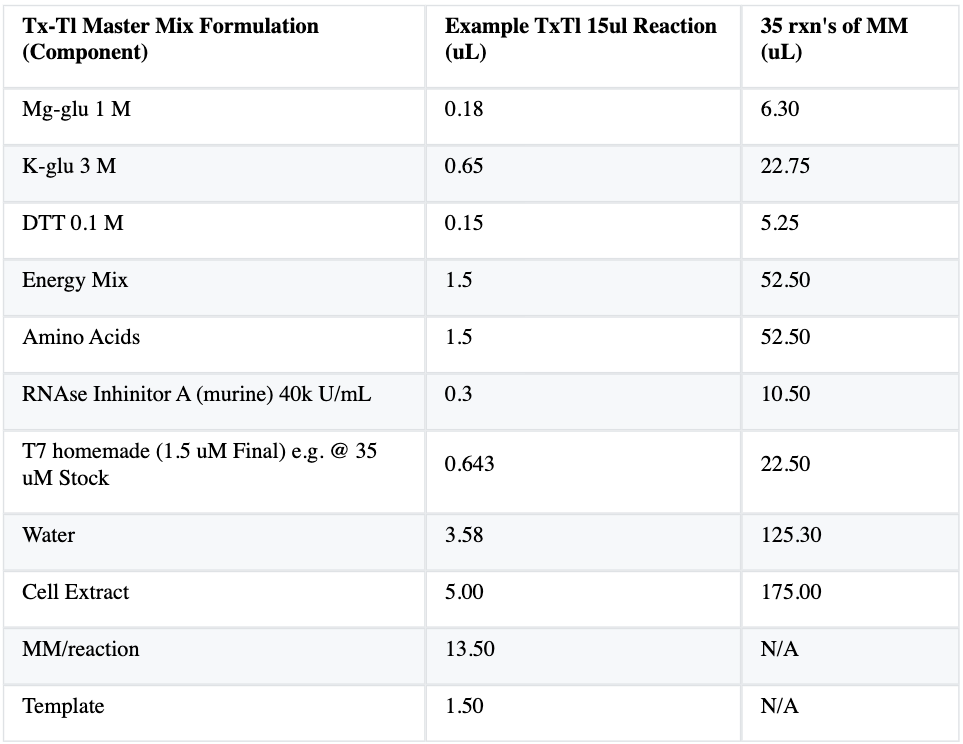
First off I will create a possitive and a negative control probes. The negative control will not have anything but water added into it. The negative control will not show any color throughout the entire process. The positive control will follow the standard procedure outlined for transcription - translation. This probe should end up with
- Negative Control - Will not be adding anything but water. - No color will be shown.
- Positive Control - Will follow standar steps by adding correct fluorscent gene. - We should see this probe turn red after the entire process has been conducted.
- Hypothesis 1 - Adding Kanamycin to the mixture - Kanamycin is an antibiotic that works by binding to a ribosomal subunit that causes misreading of mRNA. - Since this antibiotic causes a misreading of mRNA we can suppose that we will see transcription take place but not translation so the probe will be green.
- Hypothesis 2 - Adding High concentration of Puromycin to the mixture - Puromycin is an antibiotic that causes premature chain termination during translation. - This is an interesting antibiotic because it might be that we see the red gene being expressed. It depends on how prematurely translation is stopped. If probe is green we could conclude that translation was stopped before reaching the red fluorescence gene. If its red we can conclude that the gene expression was not interrupted prematurely.
- Hypothesis 3 - Adding Low concentration of Purmomycin to mixture - The results will help me understand if higher concentrations of Pouromycin make translation stop earlier.
- Hypothesis 4 - Double RNAse inhibitor - Inhibitors avoid the degradation RNA. By adding double inhibitor we would expect to still see the probe become red because the RNA will be at stable for translation to take place. We will see if there is a difference with the positive control probe. I believe their could be more expression due to the enhanced stability.
- Hypothesis 5 - No RNAse inhibitor - Since inhibitors make RNA more stable, we would expect this probe to become green and to loose its color quite quickly because of the degradation of RNA.
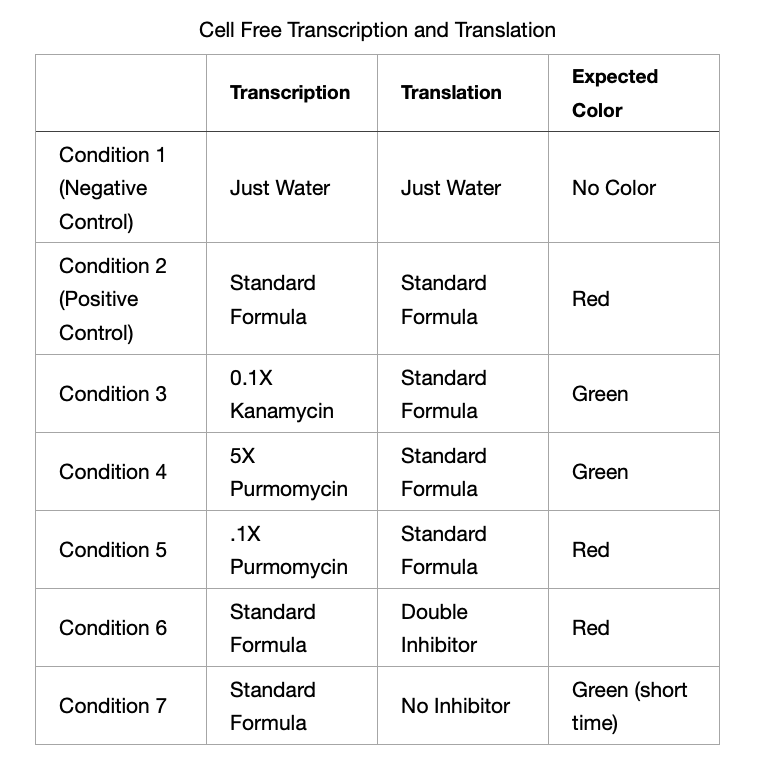
And after getting Erkin's help to perform my experiments here are my results and final experimental conditions:
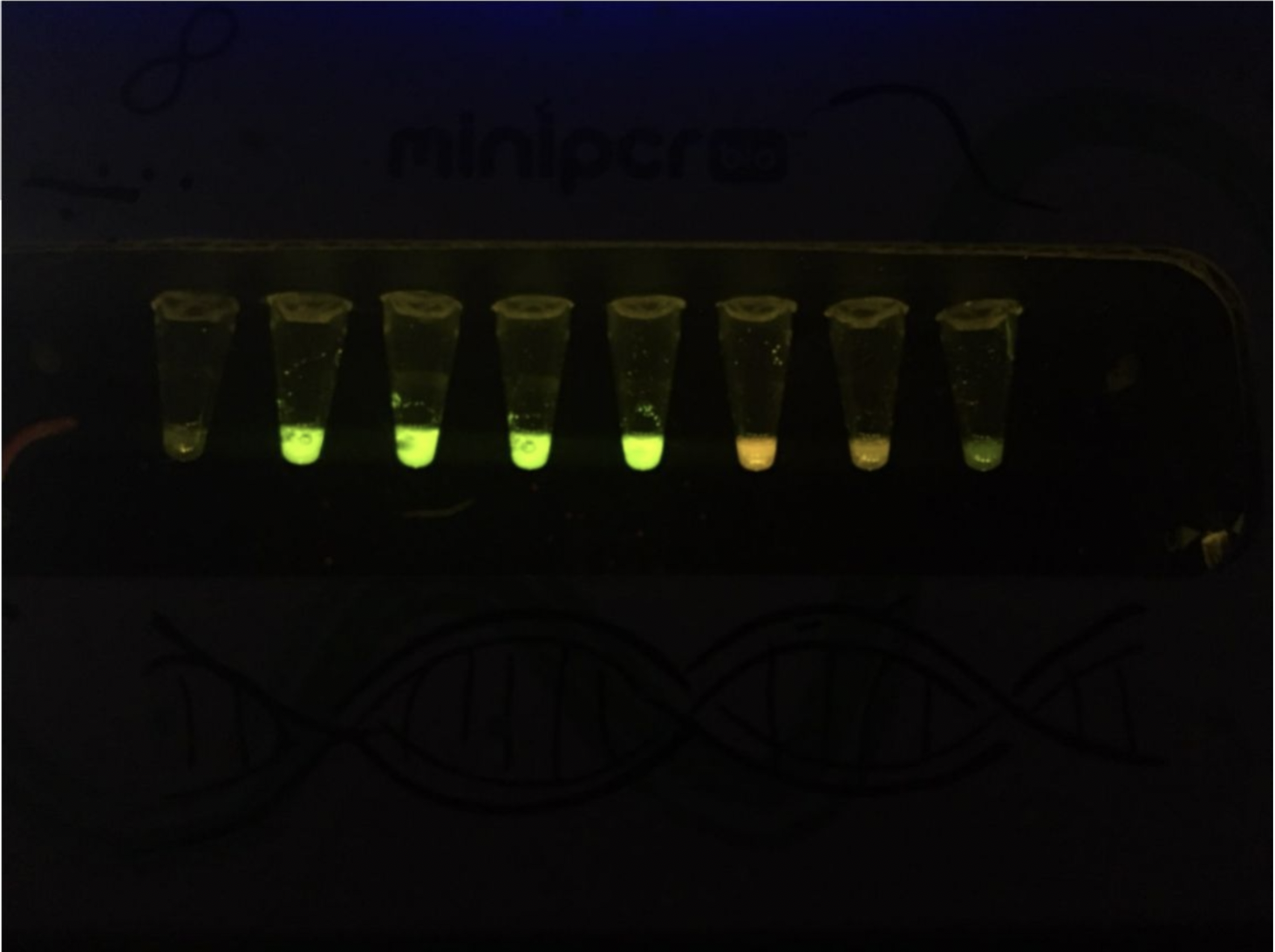

Hypothesis Check
- Tube 1 - Just Water - We see nothing as expected.
- Tube 2 - Positive control for transcription. We see green so that means that we have RNA!
- Tube 3 - Kanamycin stopped the process of translation by not allowing enxymes to binf to the RNA.
- Tube 4 - None of the tubes (4/5) expressed color red. I was expecting the one with low concentration to express but it seems that it was inhibited aas well.
- Tube 5 - None of the tubes (4/5) expressed color red. I was expecting the one with low concentration to express but it seems that it was inhibited aas well.
- Tube 6 - Posotve control for translation. We see red so it means that the formula worked and proteins were created.
- Tube 7 - RNAse maintained the RNA stable so that means that we had succesfulk translation and got proteins!
- Tube 8 - The lack of RNAse makes RNA unstable so as expected, we see no color and the color green seems to be low because RNA is not stable.
References
- [1] Jeong, Mi-Jeong, et al. "Plant gene responses to frequency-specific sound signals." Molecular breeding 21.2 (2008): 217-226.
- [2] Gu, Shaobin, Yongzhu Zhang, and Ying Wu. "Effects of sound exposure on the growth and intracellular macromolecular synthesis of E. coli k-12." PeerJ 4 (2016): e1920.
- [3] Murphy, Mark F., et al. "Acoustic vibration can enhance bacterial biofilm formation." Journal of bioscience and bioengineering 122.6 (2016): 765-770.
- [4] Acuña‐González, Edgar, David Ibarra, and Jorge Benavides. "Effects of sound elements on growth, viability and protein production yield in Escherichia coli." Journal of Chemical Technology & Biotechnology 94.4 (2019): 1100-1113.