Week 7 - Bio-production
Part A - In Silico Homework
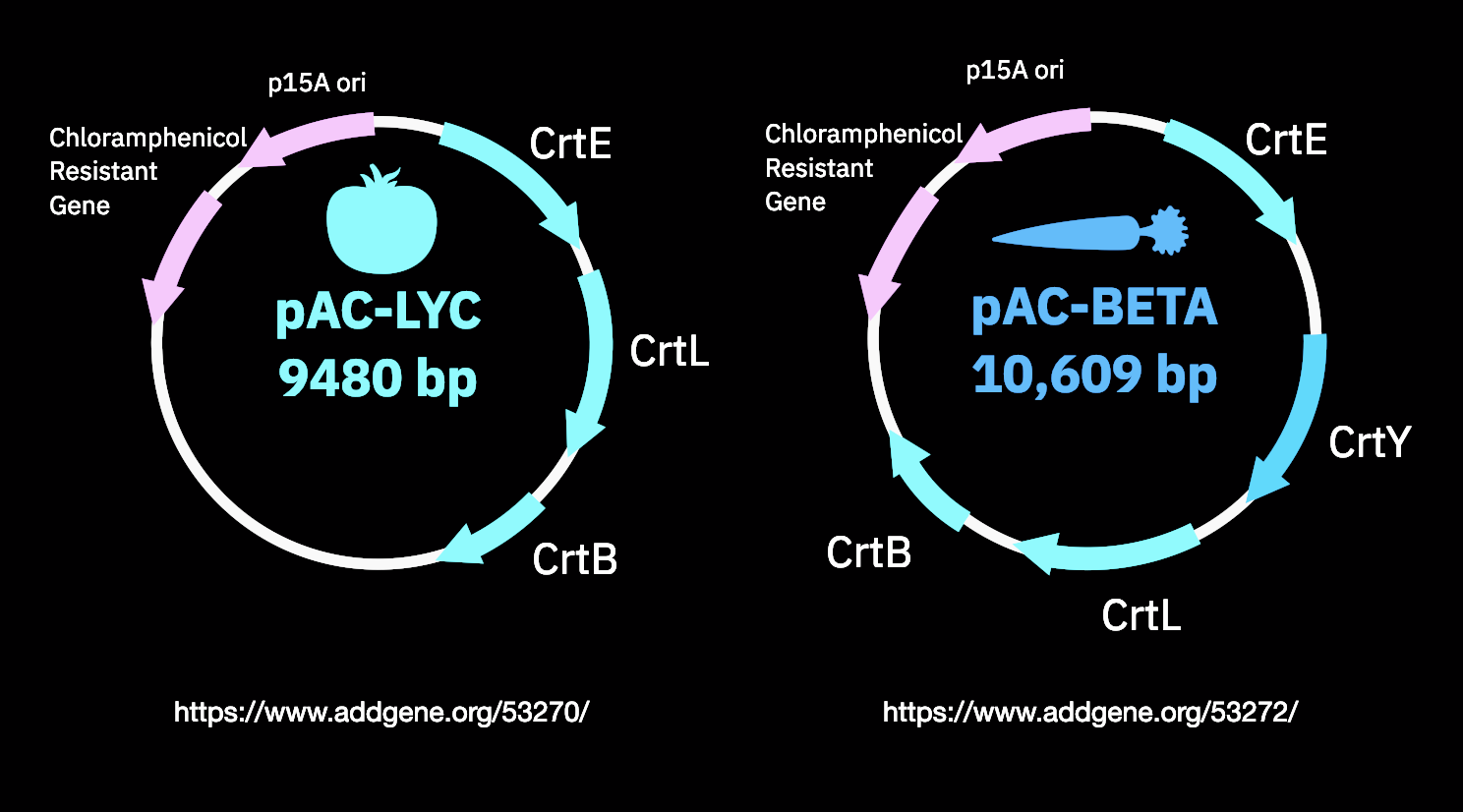
For our remote openthrons experiments this week, we will be producing Lycopene! Lycopene is a red pigment that can be found in tomatoes. We can transfer 3-enzyme pathways to E.coli to produce it in vitro. For doing so we will be using the pAC-LYC plasmid from addgene!We will also be characterizing beta-carotene production with the pAC-BETAipi plasmid. Beta-carotene is a modicifation of Lycopene that has an orange pigment. Image credits to our awesome TA for this week: Pat!
Experimental Design

As a bonus, I will be mixing the two plasmids to see if I get a mixture of colors. I also think that since the Beta is able to take Lyc as input, the final result will just be orange.
I got help from Pat to carry out my experiment remotely with the Opentrons robot! Everything seemed to work properly with the protocol. We'll see the results in a few hours!
Part B - Identifying and Ordering a Cool Gene
As part of my final project, I found out that the gene that encodes for aldolase (ald) can be regulated by stimulation at different sound frequencies.
Going through different apporaches, I realized that there is not as much work on the area as I thought but here are some of the relevant projects and genes that I found.
- Rice Plants -> Gene that Encodes for ALD + ald promoter-specific primer set + GUS reporter. [1]
- Standard E-coli + sound treatment. [2]
- E-Coli K-12 + sound treatment. [3]
- Razor Clams -> TCA Cycle -> 6-phosphofructokinase-1 - pyruvate kinase [4]
- Sound proteins -> TMC1 protein makes electrical signals when stimulated by sound. [5]
- Tomatoes -> The expression level of several ethylene biosynthetic (ACS2, ACS4, ACO1, E4 and E8) and ripening-regulated (RIN, TAGL1, HB-1, NOR, CNR) genes was influenced by sound wave treatment. [6]
- Hamster Ovary Cells -> Does not use a specific gene but a complete gene circuit that converts mechanical strain to GFP.
- AmilCP for Ecoli - This will be a recreation of the experiment we did in week 4. This will allow me to test the plasmid that I inserted into ecoli a while back. This experiment will also allow me to test the ecoli growth experiment and to test what is possible to detect from using a color sensor as a colony counter.
- The second experiment I would like to do is to test out adding the adl promoter along with the adl gene and an amilCP gene. This would let me see colors being expressed not by population but by sound stimulation.
- The third would be very similar to the second experiment but using a different gene to see if it changes anything.
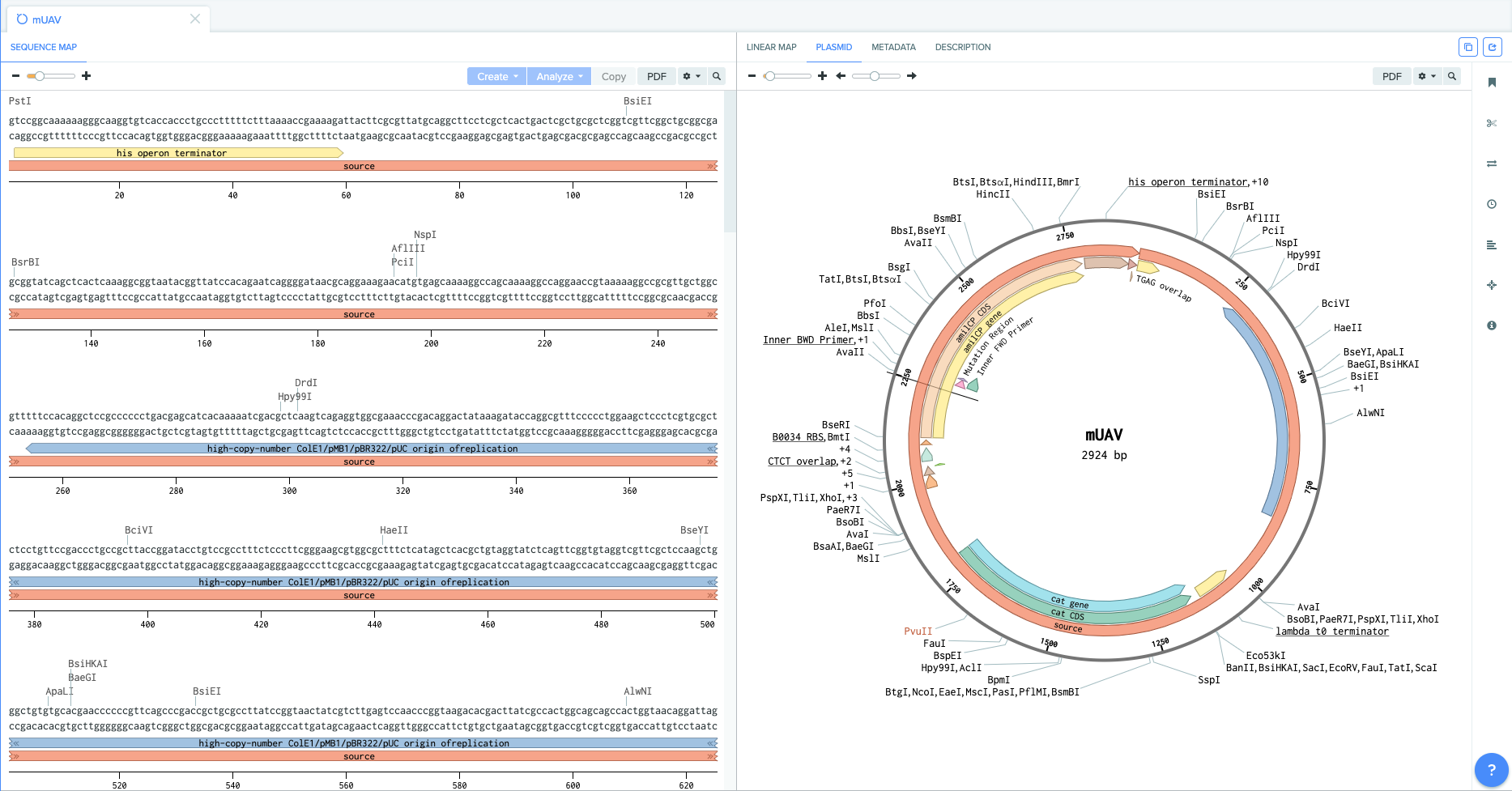
I cut out all of the origiganl backbone so that I could leave only the gene of intrest alone. I kept the regions corresponding to the genes promoter and terminator. I was left with a 872 bp segment containing the mutation region as GTCAAT (blue mutation).
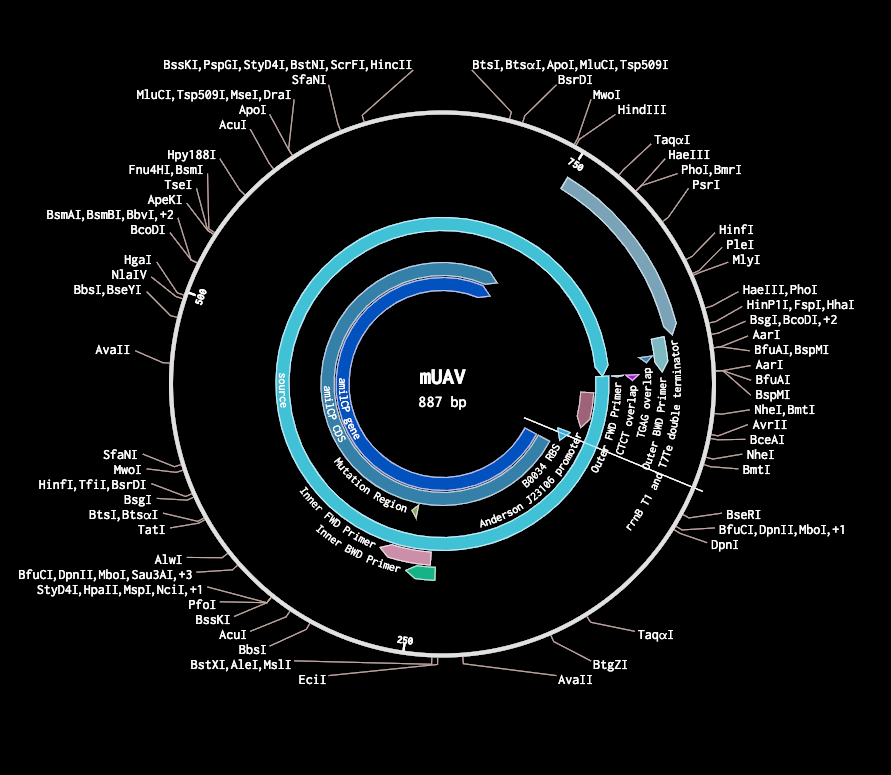
Inputing the sequence on the Twist order platform outputs no error. This means that my gene is properly made. Now, I would like to integrate it into a plasmid so that I don't have to adhere it to after it arrives and I can just input it into the e-coli.
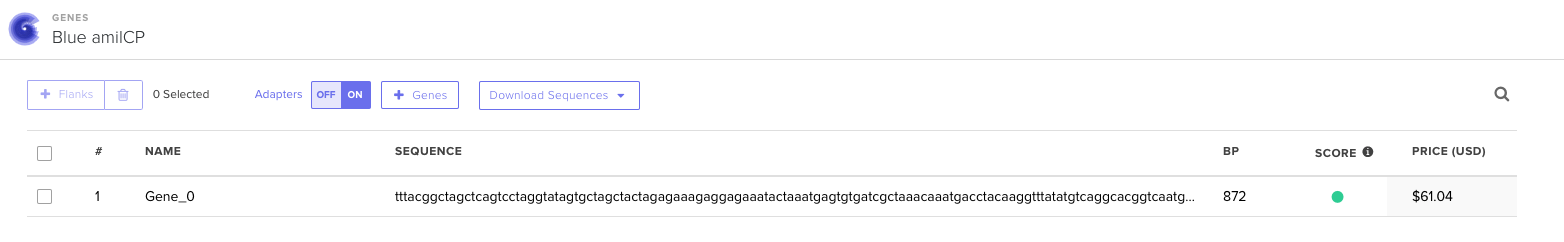
Turns out that the process I first did is the correct one for making just a gene fragment but what I really wnat and have to do is to order this fragment within a plasmid (Clonal Gene). Knowing this changes the entire process that I used previously. I then followed this steps for my second attempt:
- I copied the sequence from the mUAV plasmid that we used in week 4.
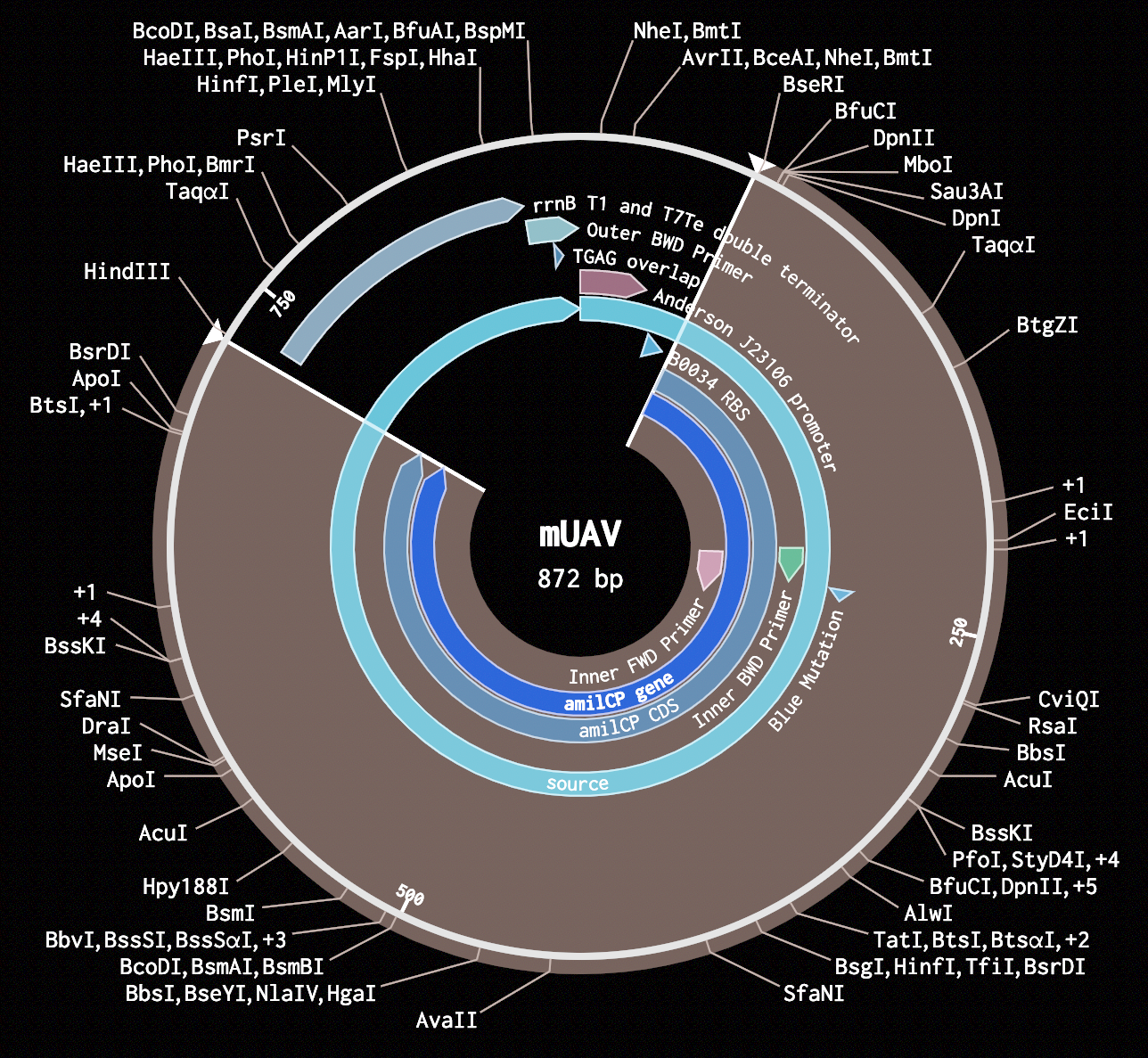
- I added the sequence corresponding to the RBS from the mUAV plasmid at the beginning of my insert.
- I chose to insert the gene inside of pet21 at EcorRI site.
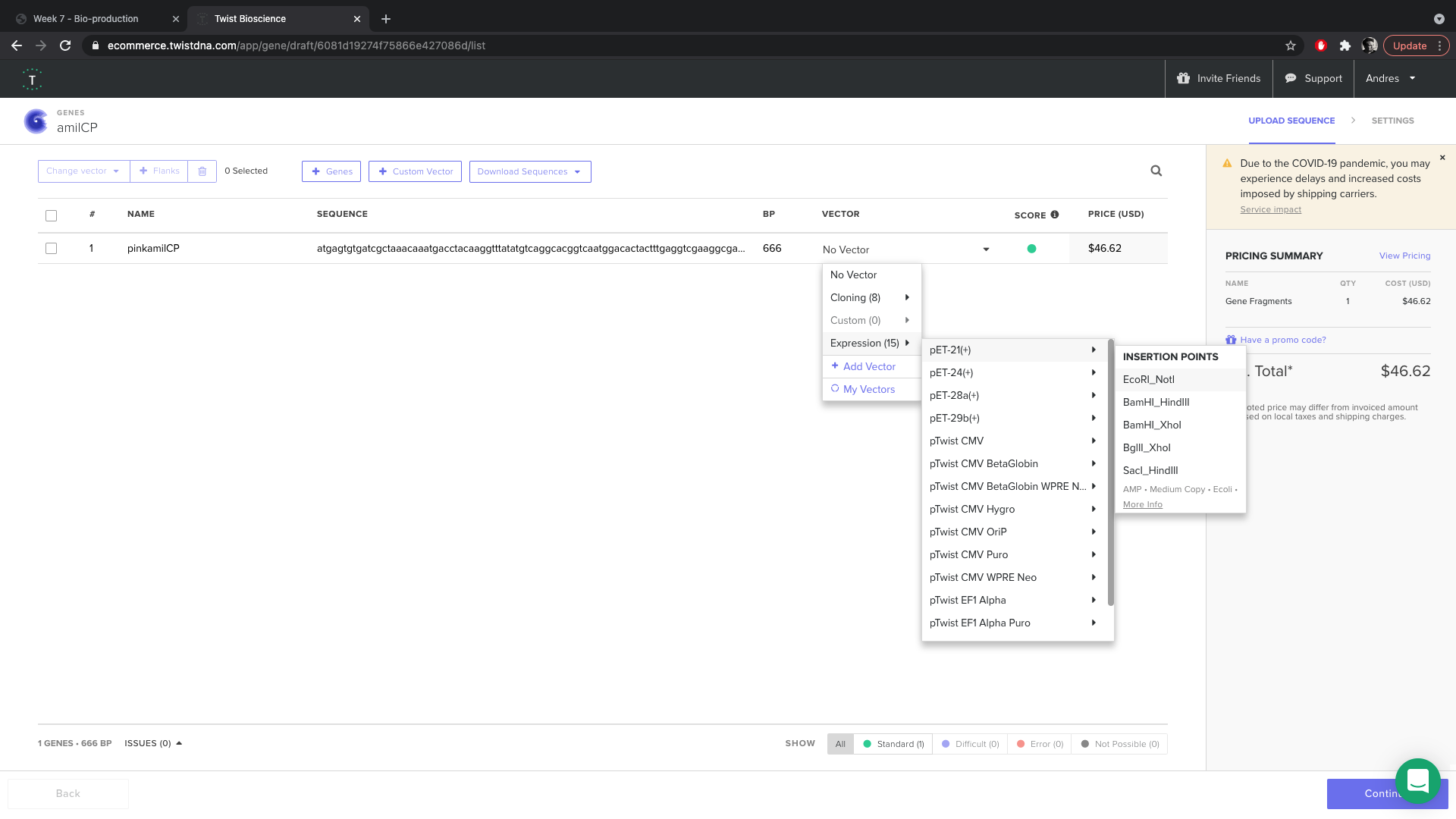
- I opened up the sequence to do codon optimization.
- At the beginning of optimization, I chose the sequence corresponding to the amil gene (leaving the sequence for the RBS unselected). It is important to make sure tat the sequence you are optimizing is divisible by 3!
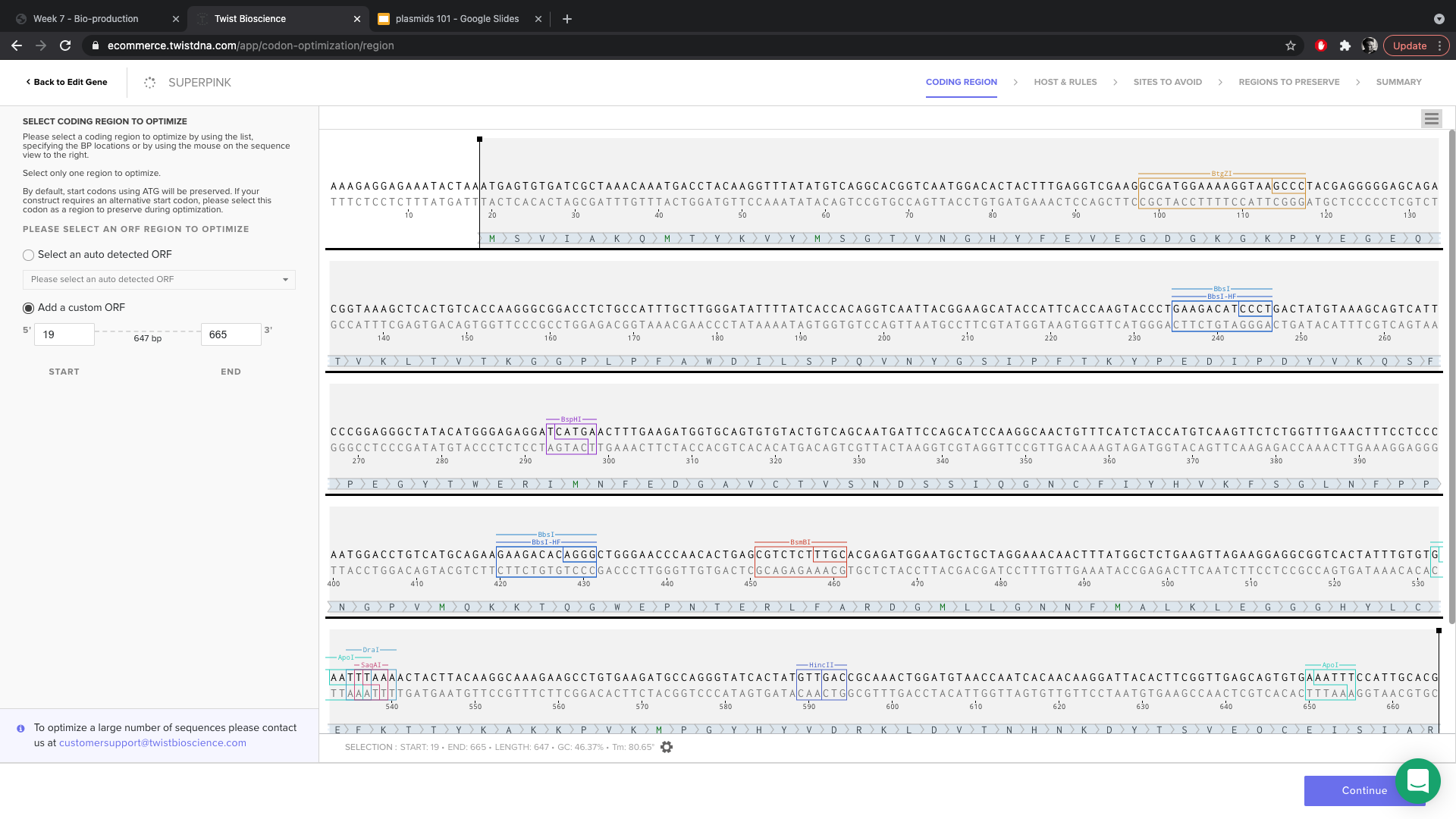
- Chose to optimize for ecoli.
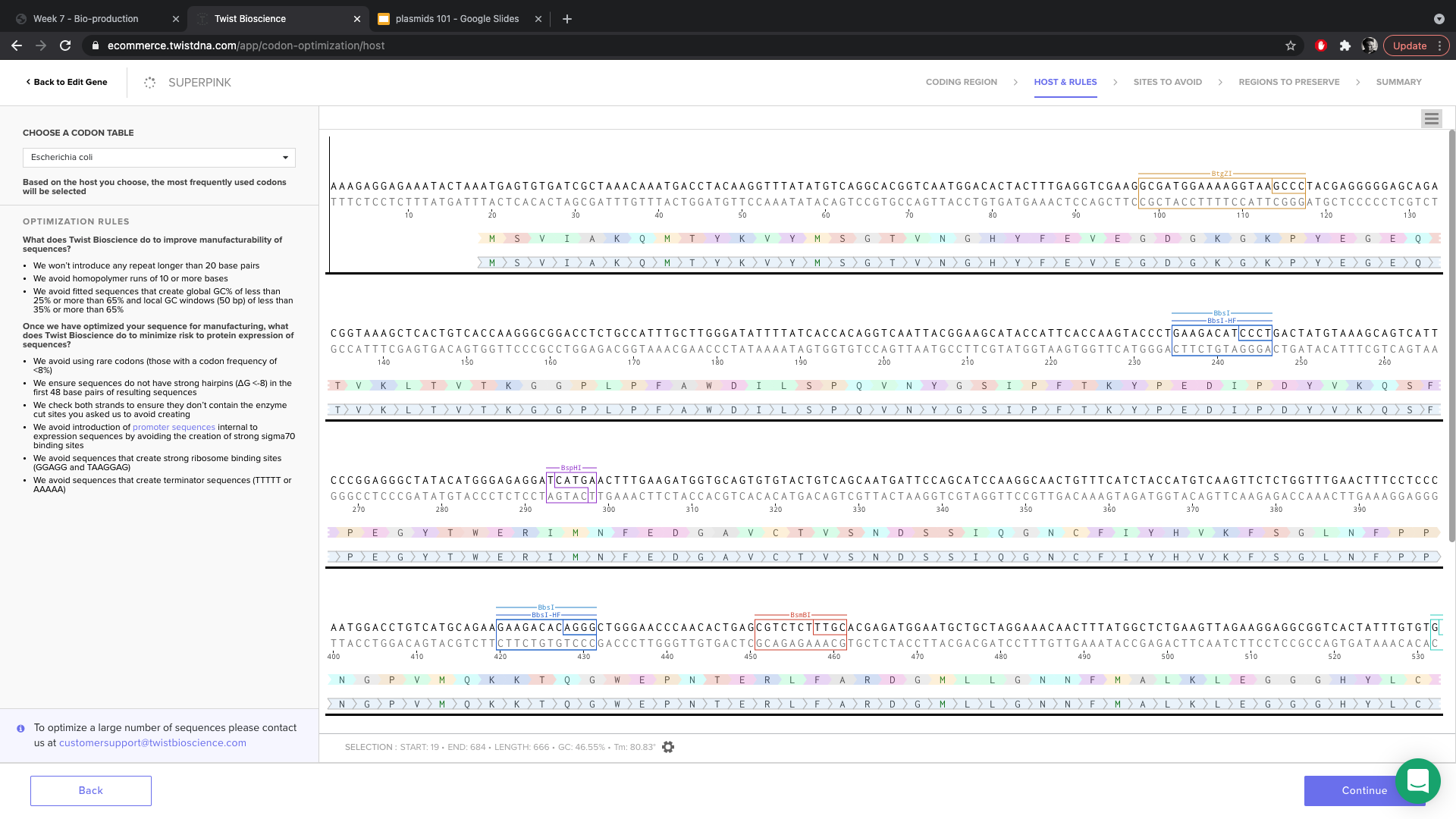
- I did not select any enzyme site to avoid(left all check boxes clear).
- When prompted about regions to preserve, I selected the region for the RBS and the region within the gene corresponding to the mutation so that we make sure that the mutation happens.
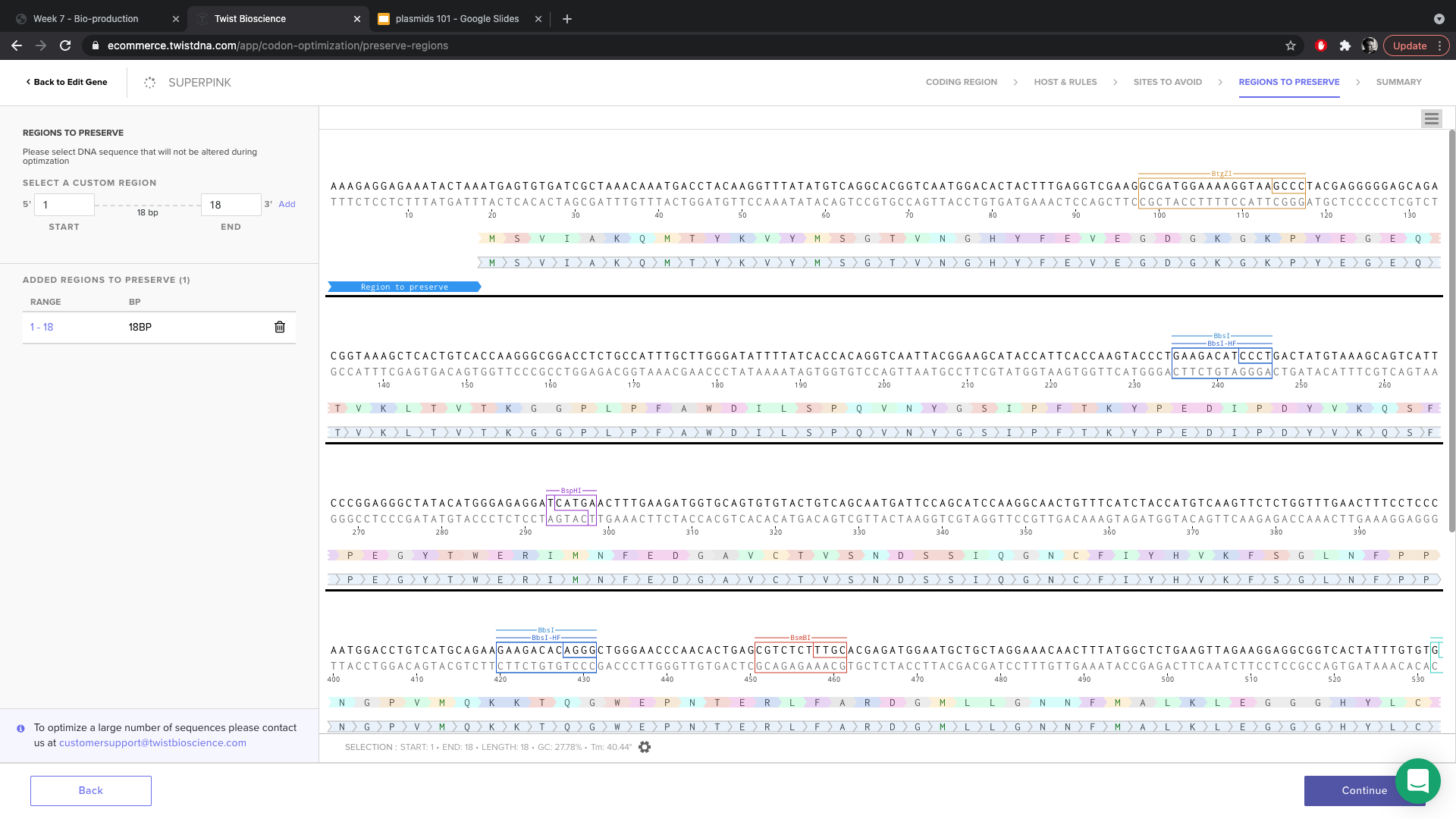
- Clicked optimize and got a nice green circle.
- Downloaded GenBank file and uploaded it to Drive.
Part C - If I had the capability of Ginko Bioworks foundry...
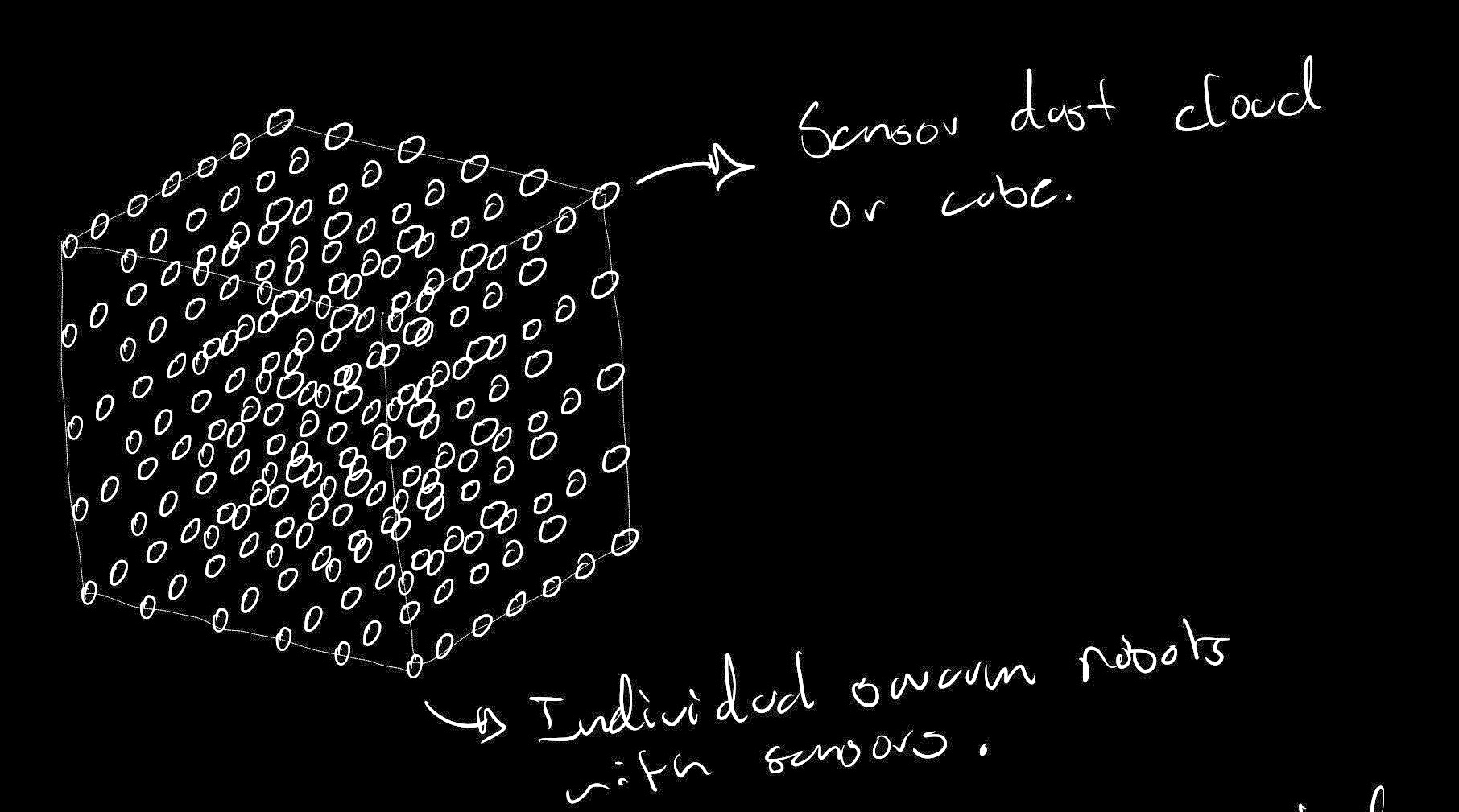
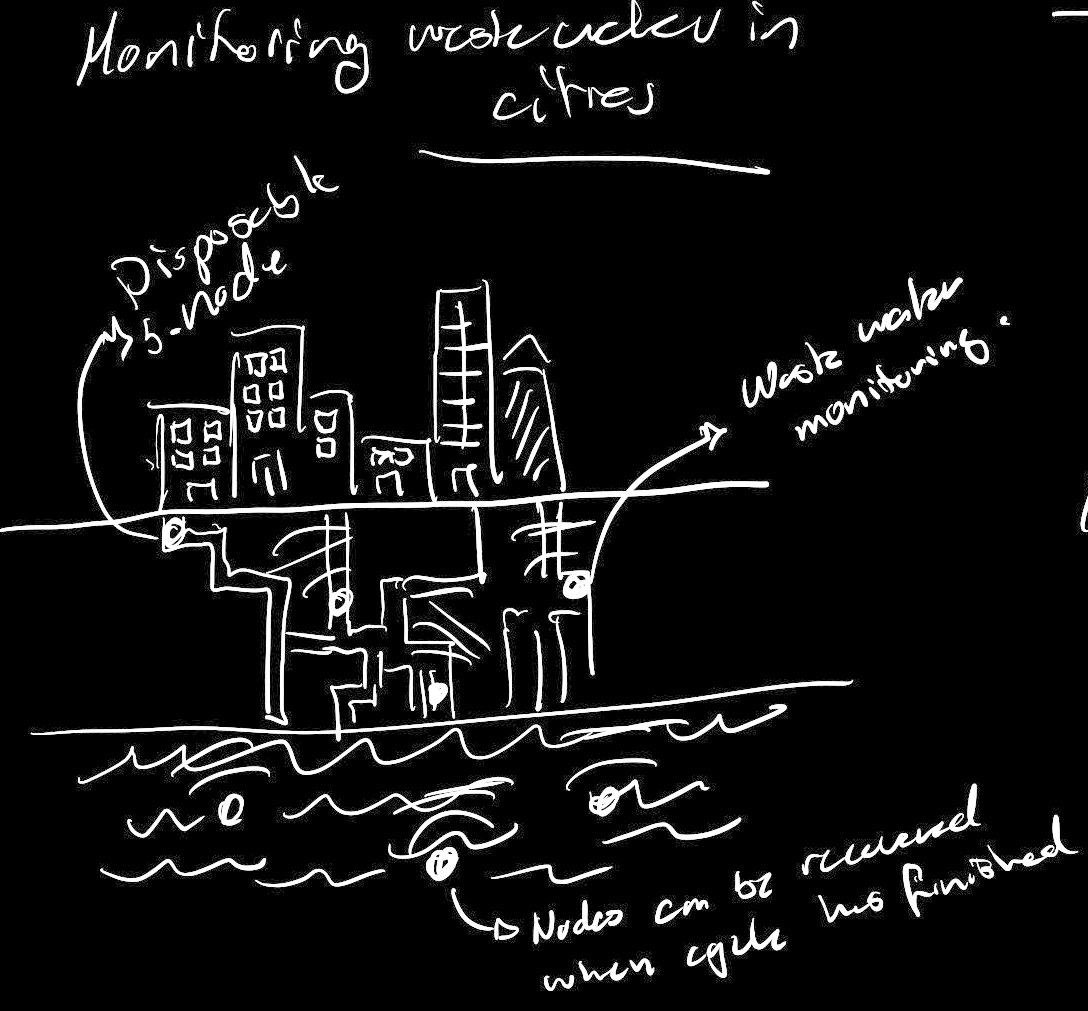
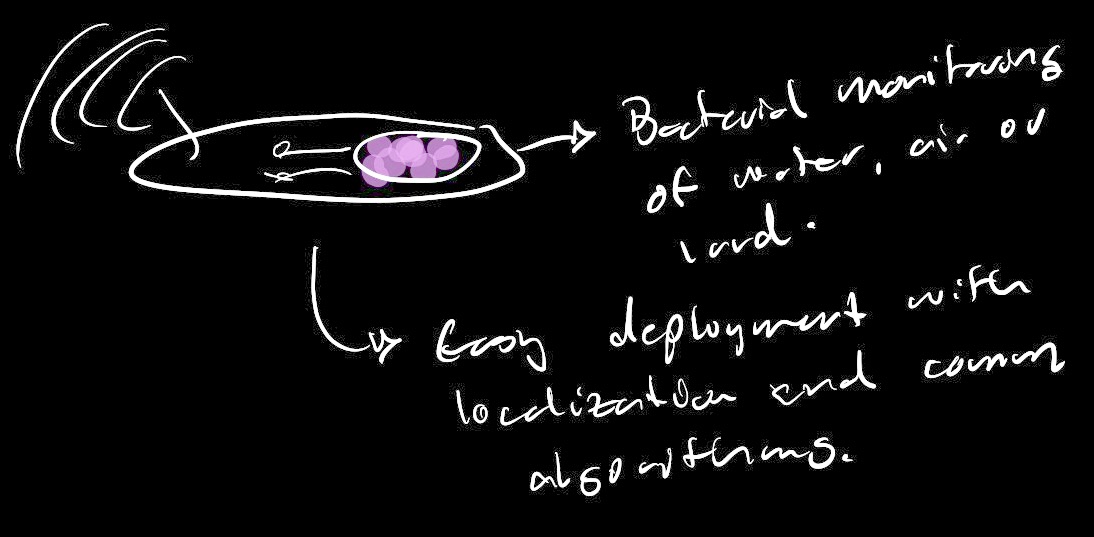
As expressed in my final project motivations, I'm very interested in what large arrays of bio sensors could mean for sensing within the context of a city. Would these networks allow us to tap into a layers of information that were previoulsy unaccessible throgh electromechanical sensing? Maybe the networks Would allow us to gain better health information (pollution, diseases, radiaton). Maybe they could be used for understanding the complex interactions that cities have with the natural environment (Human activity impacting ecosystems). Another interesting idea would be to use them as a means to better understand social and human behavior. Maybe some of the enzymes that govern our social behavior could be detected within public spaces so that we are able to understand community engagement and interactions.
With this in mind, I would see this systems as cell free or even freeze dried solutions that float around our cities like sensor dust. This would mimic the dynamics used by nature in essential processes such as pollen redistribution. Information, can float around the city and arrive to specific receptors that allow us to understand the history that a specific particle has had. Ginko's foundries would allow us to crete this dust bio particles in a way that can be scaled to the number of sensor nodes that would be needed for an approach like this to work. Also, we could use some of the techniques discussed in class to make this sensor safe when interacting with the living environment. Nodes would serve their functional life and then be unreactive to nature until they degrade.