The human gut microbiota is one of the most densely populated ecosystems of microorganisms on earth. With an estimated 100 trillion microorganisms, the gut is an extraordinarily complex system of microbe-microbe and microbe-host interactions. A growing body of research is beginning to elucidate the diverse impacts the gut microbiota plays in human health and development, from nutrition, to disease, and even cognition. Recently, with the success of fecal matter transplants (FMTs) to treat infectious disease, microbes are emerging as a unique therapeutic. Model systems to both prototype and study complex polymicrobial systems are a necessity for producing robust microbial communities that can be engineered at both the genetic level (subcellular) and population level (multicellular).
Part A: Isolate a colony from your skin and mouth and identify them
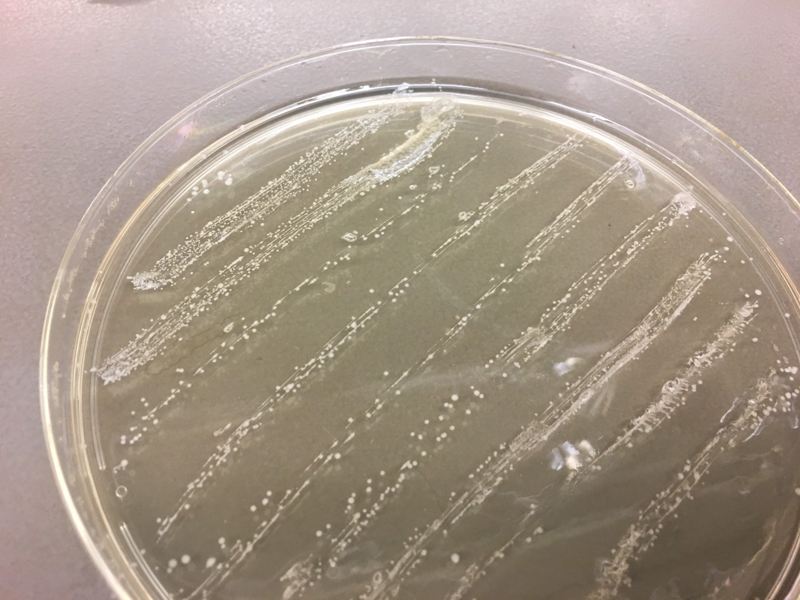
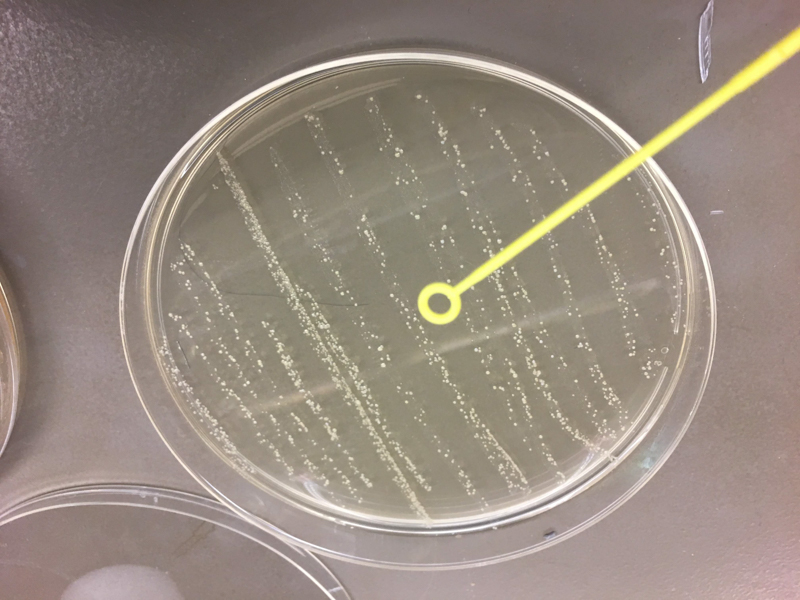
We used cotton swabs to transfer our skin and oral microbiome to streak LB+agar petri dish. After labeling the plates, we incubated them overnight at 37°C. Next, we picked the colonies to sequence, isolated them and restreaked them on a new LB+agar plate and incubated again the samples at 37°C. Then, we sent them to Genewiz for 16s rrna sequencing. Once we received the result we used Targeted Loci Nucleotide BLAST to blast our own sequence and compared them to the result provided by Genewiz.
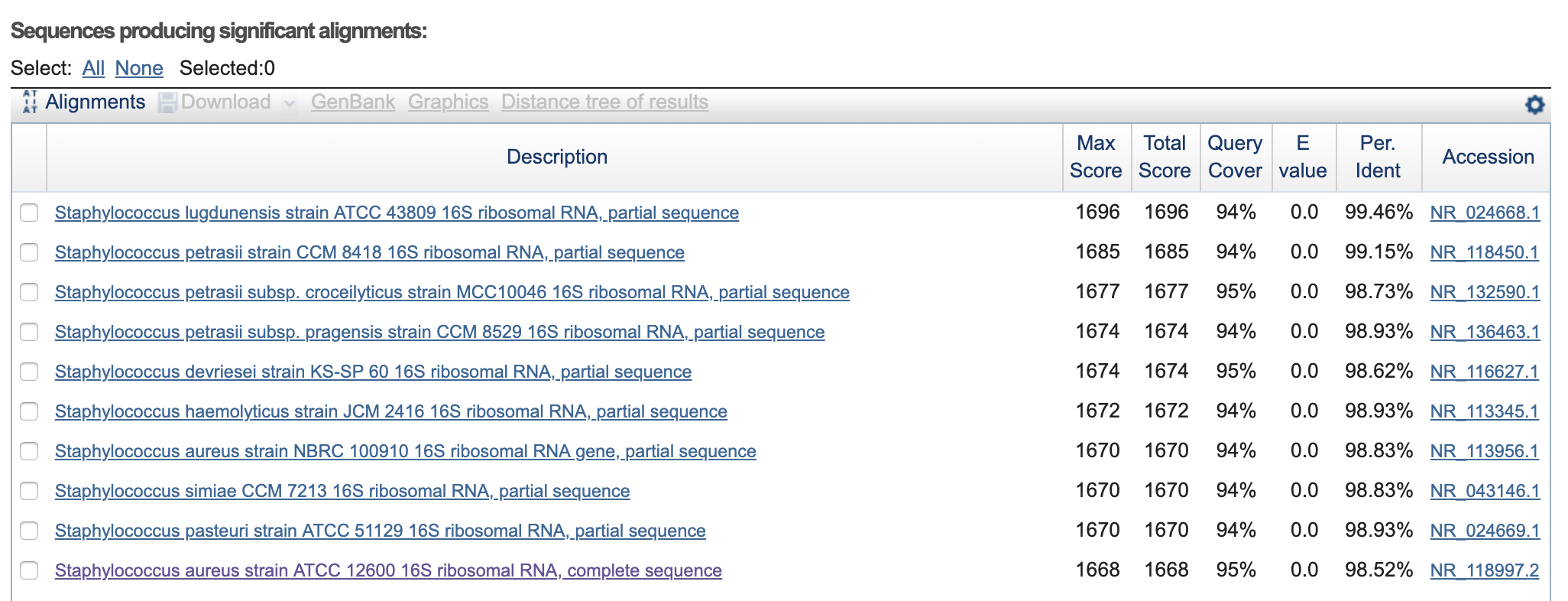
Oscar_Mouth-16s-rRNA-SeqF.seq >>> Staphylococcus aureus strain ATCC 12600 16S ribosomal RNA, complete sequence.
Oscar_Mouth-16s-rRNA-SeqR.seq >>> Staphylococcus aureus strain S33 R 16S ribosomal RNA, complete sequence.
Oscar_Skin-16S-rRNA-seqF.seq >>> Staphylococcus lugdunensis strain ATCC 43809 16S ribosomal RNA, partial sequence.
Oscar_Skin-16S-rRNA-seqR.seq >>> Staphylococcus lugdunensis strain ATCC 43809 16S ribosomal RNA, partial sequence
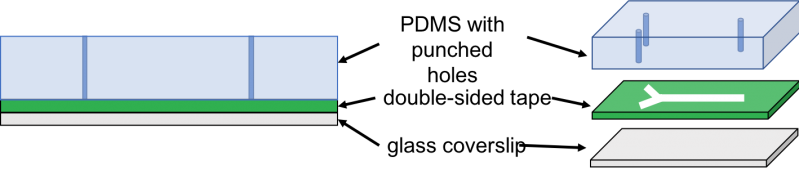
Part B: Make a tape based microfluidic device to confine bacteria and image them using a microscope
We cured PDMS on a petri dish and laser-cut double sided kapton tape. Then, we used a pen knife to cut a slab of PDMS of the size of laser-cut tape. Next, we used tweezers to peel off one side of the tape and adhere the exposed side to the PDMS slab. Then, we made holes through the PDMS by punching the inlets and outlet of the lasercut tape using a biopsy puncher. Finally, we peeled off the other side of the tape and adhere it to the coverslip or glass slide. Then, we cut three tubes of about 10cm each and inserted them in the inlets and outlets. Next, we used the syringes to inject the cell culture into the inlet tubing and imaged the result under 40x and 60x magnification.
Part C: Make your own fluidic device, via tape, 3D printing, or another method
We designed and built our own microfluidic petri dish designed to activate optogenetically modified bacteria. The device is desinged using Rhinoceros 3d CAD software and printed using the Formlabs 2 3D printer. Our device can be programmed to activate the bacteria at specified time intervals and with target light frequencies. We envision our device to be a first step towards wearable microfluidics.