Engineering Gut Microbiome
Find a research or journal article where researchers cultivate 2 or more microbial strains. What technology are they utilizing? How scalable is this approach to more than 2 strains? How do they address issues related to requiring multiple media?
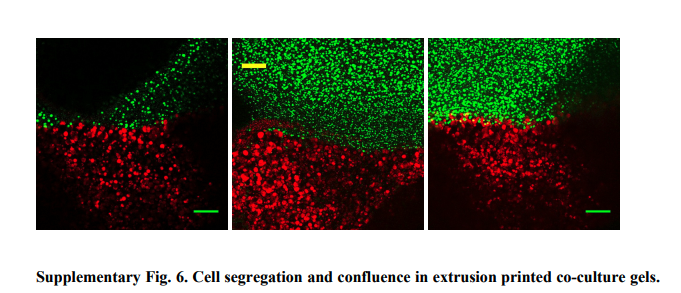
I looked at Compartmentalized microbes and co-cultures in hydrogels for on-demand bioproduction and preservation
They demonstrate a platform that utilizes a hydrogel system to compartmentalize microbes to build spatially segregated microbial consortia.
They show that co-culture systems immobilized within hydrogels can be extrusion printed to form solid-state bioreactors capable of producing small molecules and antimicrobial peptides for multiple, repeated cycles of use.
They also demonstrate the microbe-laden hydrogels can also be preserved via lyophilization, stored in a dried state, and then rehydrated at a later time for on-demand chemical production.
Propose a technology for culturing 2 or more strains. How might you innovate in this area given the paper you reviewed?
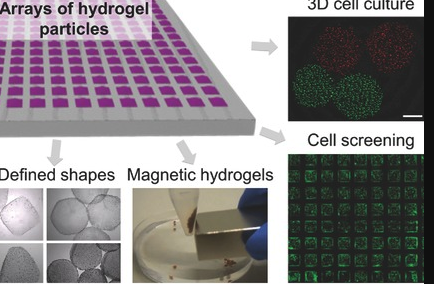
I suggest using microarray hydrogel patterning to increase the number of samples. And find better surface specific binding affinity for each individual cell-type based on chemical weight or other biochemical properties. For co-culturing known stranging, could possible to have a barcode for each individual spatial location of hydrogel cells.
One of the great challenges in microbiology currently is culturing “unculturable” microbes. Propose a methodology for how you might explore this significant space of uncultured microbes.
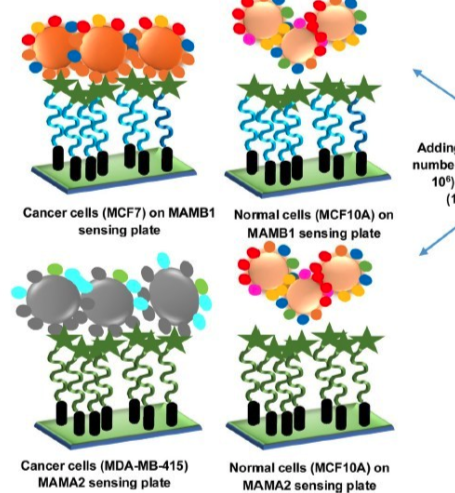
Review an article on an artificial gut-on-a-chip technology. What scientific hypotheses are they testing with this in vitro tool? Could you propose an upgrade or innovation to their technique to enable the exploration of other scientific hypotheses? Provide an example of at least one hypothesis you would explore with your proposed system.
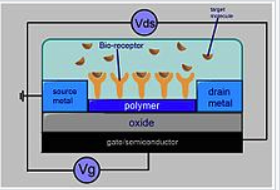
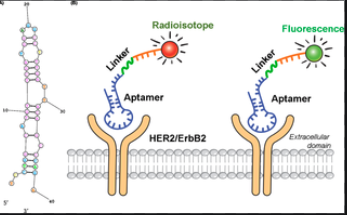
We could measure the change the voltage of based on specific Aptamer Binding. If they bind to specific unknown strains,then we could figure out if there are apratmer binding.
One of the biggest challenges in public health is quickly detecting new disease outbreaks. How would you go about adaptively responding to new outbreaks? Biobots is using qPCR, so you need to know what you are looking for. How might you develop a technology with a more general view? Some example approaches include microfluidics and point-of-care sequencing, but what else? In particular, are there ways to look for RNA viruses like the flu and SARS-CoV2?
