METHODOLOGY & RESULTS
BEE COLLECTION
We collected and removed the stingers from 12 bees.
Bees were stored overnight in the fridge in a petri dish.

insects are interesting, but so weird up close.
CULTURING THE BEE MICROBIOME
We tried a few different methods of sampling the bee’s microbiome, summarized below:
Swab the bee’s fur and head with a clean q-tip, then swab onto plate.
Roll the entire bee around on plate.
Cut upon the bee between the head and abdomen. Swab the inner goo with a clean q-tip, then swab onto plate.
Some black liquid oozed out of the abdomen compartment. Used q-tip to transfer the liquid to plate, though q-tip absorbed much of the liquid.
As above, except diluted the black liquid with water and pipetted the liquid onto plate, spreading with pipette tip.

BEE POOP. BEEE POOOOP.
" diarrhea? diaBEEea? " - Vivian
trying to identify intestinal structure but it's just goo.
We used YPD plates containing 10% dextrose and 50 μg/mL chloramphenicol (medium A) to inhibit the growth of filamentous fungi and bacteria, respectively.
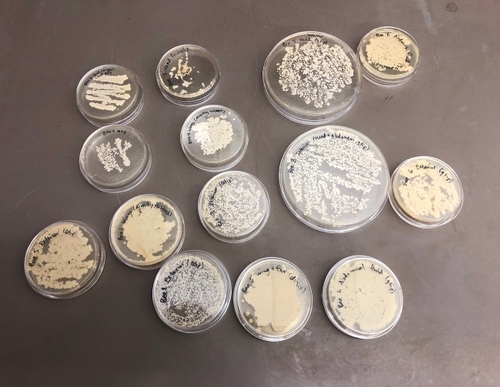
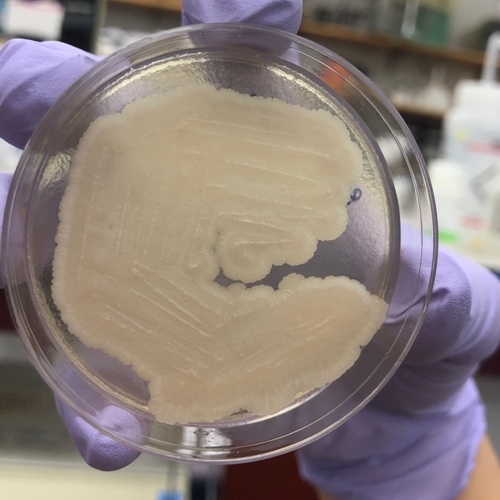
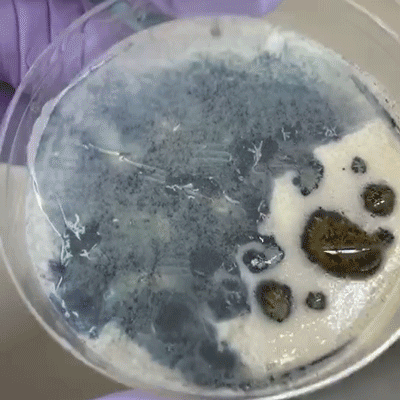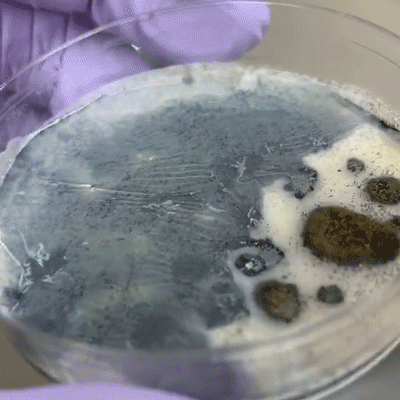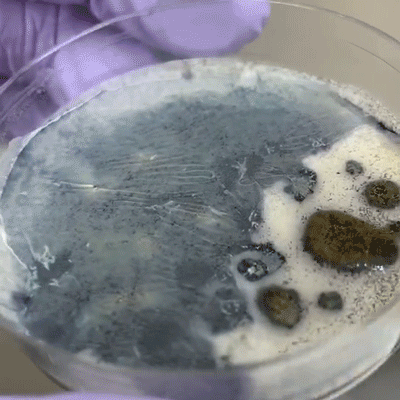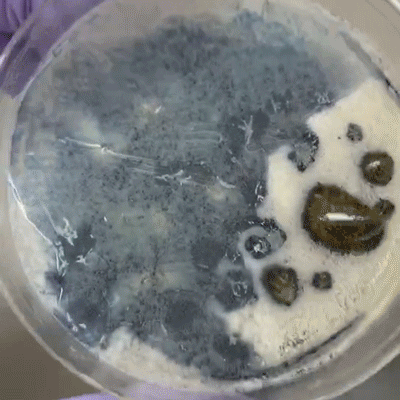
We incubated the plates at 30C for two days, after which we observed growth on all plates, all of which smelled like yeast as we know it from baker’s yeast. There was no obvious bacterial or mold contamination.
ISOLATING INDIVIDUAL YEAST
Most of the plates had multiple colors and/or morphologies of microbes. We used pipette tips to sample as many different, unique microbes as we could determine from color and morphology, and grew the samples on new selective YPD plates (Reagent A). In order to get single colonies (a mass of genetically-identical microbes that divided from a single microbe), we used the streaking technique. Incubation at 30C for 24 hours was sufficient for most of the microbes to grow into decently-sized colonies; the pink yeast showed slower growth.
We selected 14 strains for further analysis, with the expectation that there are at least 6 unique strains.
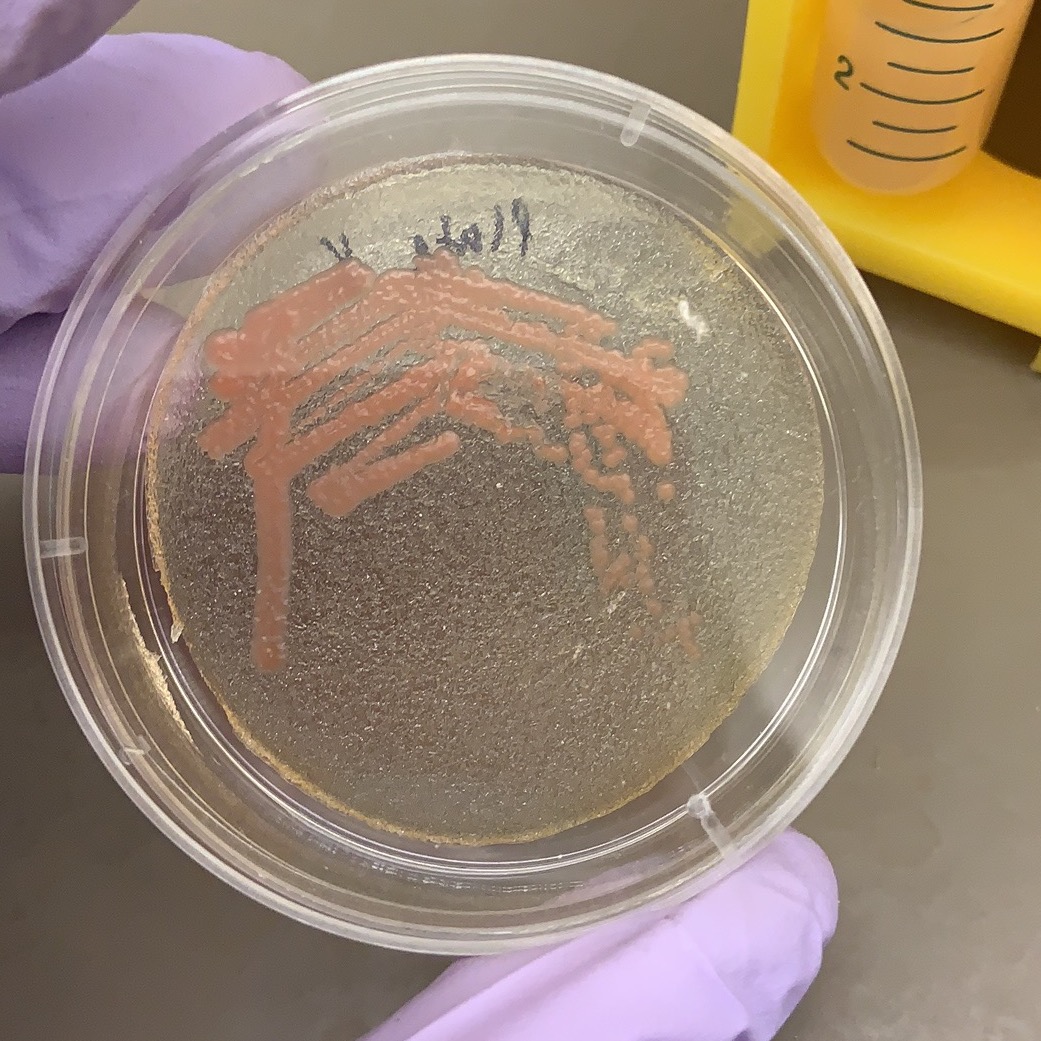
A few of the isolated yeasts shared a similar color and morphology to yeast of the Rhodotorula genus, which have been isolated from traditional breads.

TESTING FOR AMYLASE (STARCH DEGRADATION)
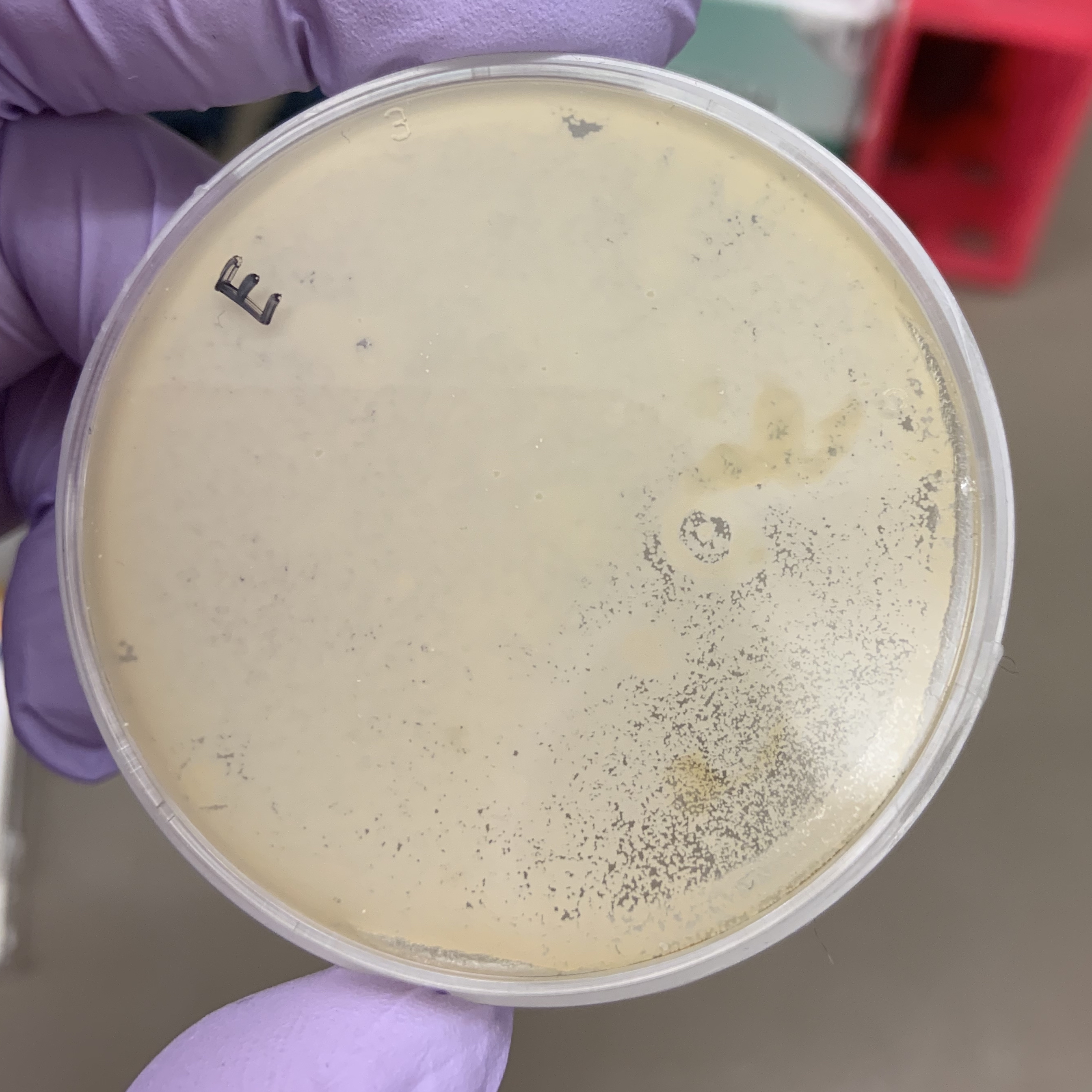
In order to test for the expression of amylase in the yeast strains, we streaked colonies onto YPS (yeast extract, peptone, and starch) plates and incubated at 30C overnight. We then stained the plates with iodine to identify areas of starch degradation into sugars, an indication that the yeast expresses amylase — starch turns blue-black when reacting with iodine. Unfortunately, none of the yeast colonies had “halos” of white around them.
TESTING FOR CO2 PRODUCTION
We first attempted to test for CO2 production by culturing the yeast colonies in liquid YPD broth and securing balloons over the mouths of the culture tubes, then incubating on a spinner at 30C. Yeast is a microscopic fungus that converts sugar into carbon dioxide. The generated CO2 gas should fill the tube and then fill the balloon as more gas is created. There was no obvious inflation of the balloon; however, because the positive control, baker’s yeast, also did not yield inflation of the balloon, it is possible the methodology was faulty.
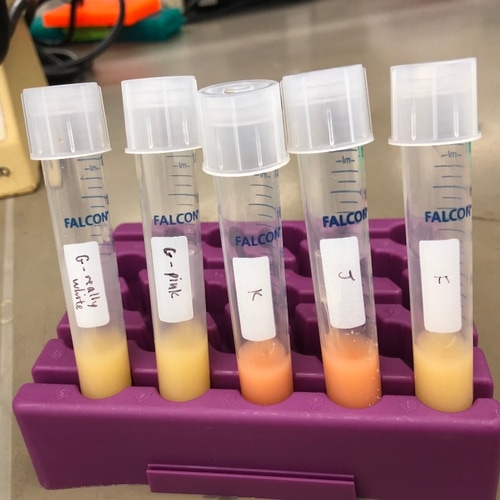
Since dissolved CO2 would decrease the pH of the medium, we also tested for pH. Most of the yeasts were of pH 7-7.5, but one of the pink yeasts had pH ~6, while another had pH <5. However, it is possible that metabolites secreted by the yeast resulted in the decrease in pH.
IODINE ( AND THE STRANGE EFFECT ?)
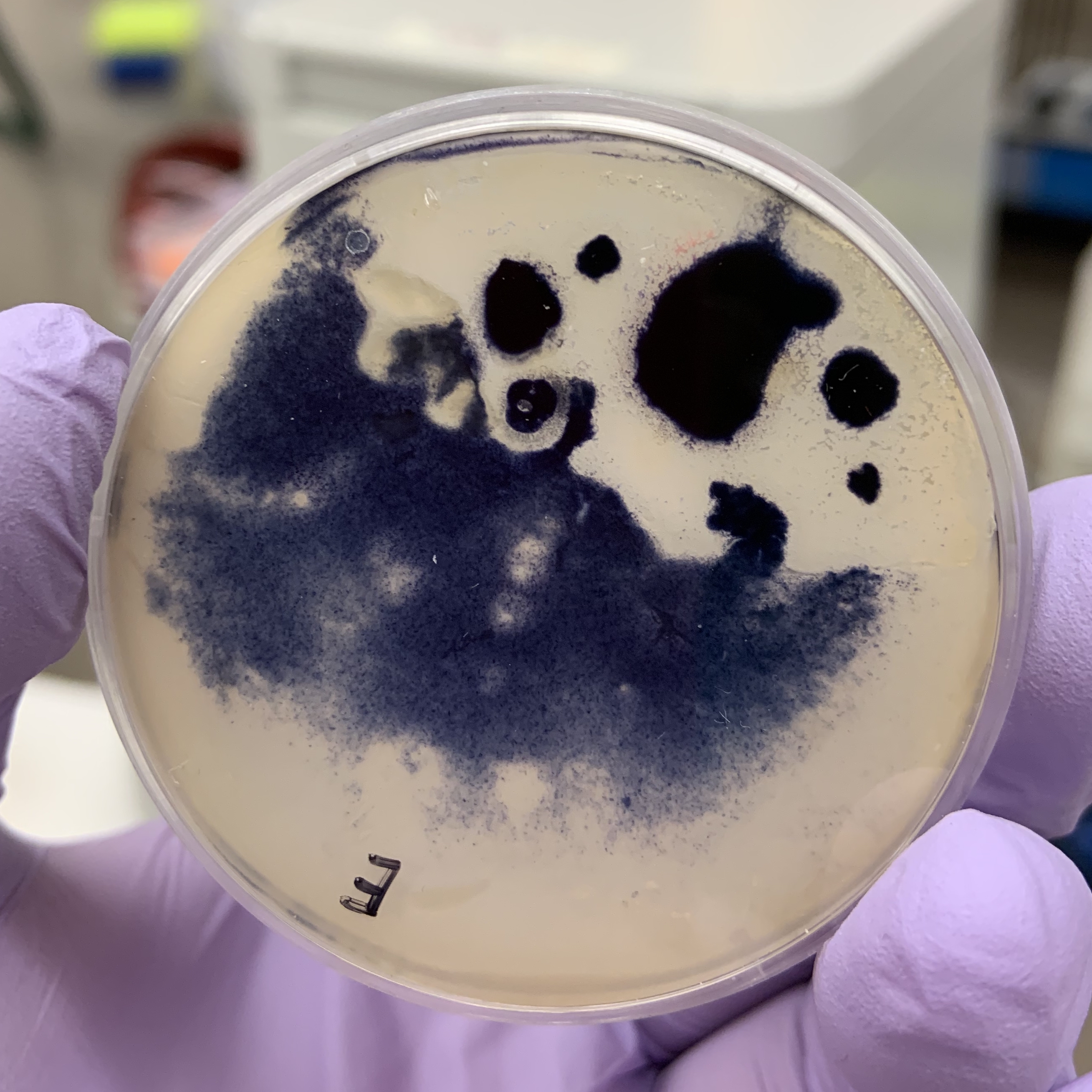
With starch added to the agar medium we can visually see the development of the yeast. Iodine was added to the plate after incubation to detect if the yeast has hydrolyze starch. The solution turns blue, purple, or black (depending on the concentration of iodine) in the presence of starch. A clearing around the bacterial growth indicates that the organism has hydrolyzed starch.
A strange observation was that the yeast sample somehow grew a layer of penetrable thin membrane, that when we pipetted Iodine onto its surface, the liquid seeps through and delaminates the thin membrane from the bottom layer, and flows around underneath the membrane.
SEQUENCING
We will be sending 14 strains out for 26S rRNA sequencing (like 16S, except fungi have a 26S ribosome subunit instead of bacteria’s 16S) to hopefully identify the yeast we isolated (perhaps there’s a new strain!) and screen for pathogenicity. To conduct a more thorough analysis on whether or not the yeast on our hand are safe to use in food, we would have to conduct metagenomics sequencing and screen for known genes for toxic metabolites and/or human pathogenicity.
< CONSIDERATIONS & ETHICS OF BIOLOGY>
BEE COMMUNITY
Bees are much more important than what we usually thought.
What’s often perceived as species only beneficial to the environment can still have incredible relevance to human, not only to our understanding of greater ecology, but in addition surprising commercial production and industrial values. Obtaining and utilizing the uniqueness of these yeasts requires maintaining the uniqueness of the these bees’ microbial communities.
It raises a question of how much control and manipulation we should have of the right to do when it comes to these populations as an ethical concern of biology. Is there a method we can use to safety obtain microbial data and yeast samples from the bees without interrupting their local communities?
HUMAN FACTOR AS AN ADVANTAGE
By involving human in the process, we can start implementing procedures to help advance the current bee keeping and honey production procedures and set up. For example on toxins, virus and antibiotics control.
Bee products (honey) are polluted via different sources of contaminants including pesticides, bacteria, PCBs, antibiotics, etc. As these product are widely consumed by people, their contamination may carry serious health hazards and risks. Pesticide residues cause genetic mutations and cellular degradation and presence of antibiotics might increase resistant human or animal pathogens.
If bee yeasts are used more frequently in commercial food production, there will likely be more proper measurements taken into considering when maintaining a healthy beehive or administrating drugs / antibiotics to bees for food safety reasons.
MATERIALS
Medium A: YPD agar, selective for yeast
Components (for 1L):
- 65g YPD agar powder (Sigma-Aldrich), containing yeast extract, peptone, and agar
- 100g dextrose (Sigma-Aldrich)
Protocol:
- Dissolve the dextrose thoroughly in water first, using the microwave cautiously to encourage dissolving without burning the sugar.
- Add the YPD agar powder, and heat in microwave, swirling bottle every minute, till just boiling. If you didn’t dissolve the dextrose thoroughly in water, it will likely caramelize/burn long before the agar dissolves.
- Once the bottle is cool enough to touch, add the chloramphenicol, swirl to mix, and plate.
Medium B: YPS agar
Components (for 1L):
- 1L YP broth (Bio-world)
- 15g Bacto agar (Carolina)
- 10g soluble starch (Sigma-Aldrich)
Protocol:
- Dissolve starch fully in broth
- Dissolve agar fully in brothIodine (Sigma-Aldrich)
FUTURE WORKS
There are multiple things that we can do afterwards for the project.
This project can be replicated with many organisms, including more easily acquired specimens such as fruit and veggies. It is a good way to get a community engaged in biology, as well as to contribute to re-diversifying bread production.
Side note: an AI recognition software to identify yeast based on color and morphology would be a great tool for a community effort.
OTHER OBSERVATIONS
- All strains grew perceptibly more slowly on LB agar than on YPD, but two strains in particular that were isolated from the exterior of the one of the bees had almost no growth on LB.
- Yeast that are putatively the same strains (based on color and morphology) could be found both on the exterior and interior of the bee