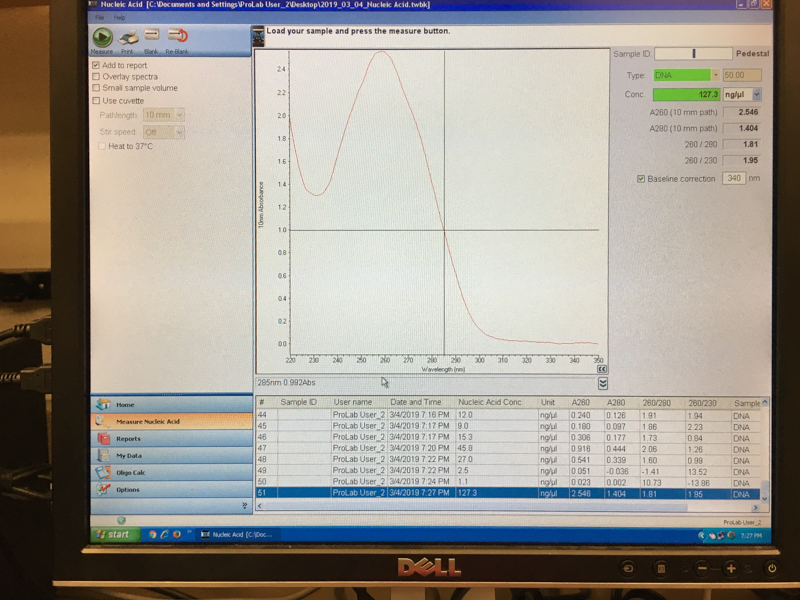
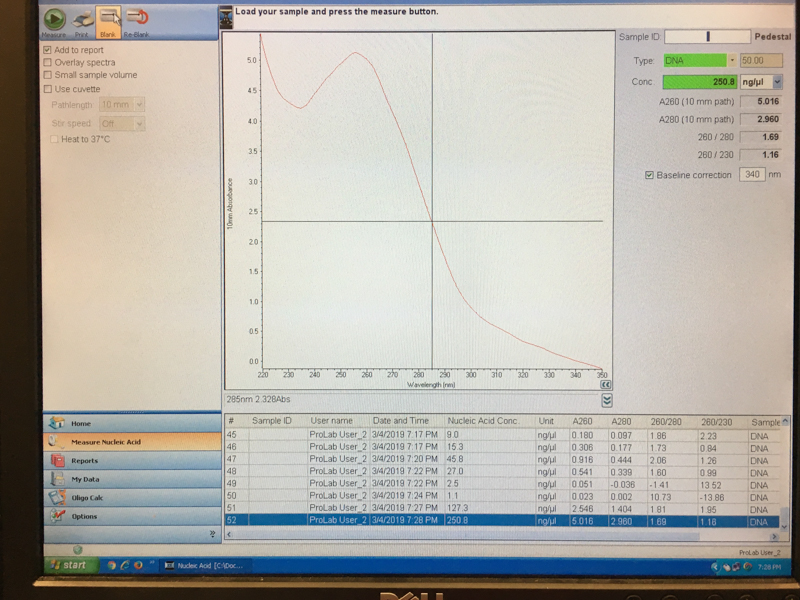
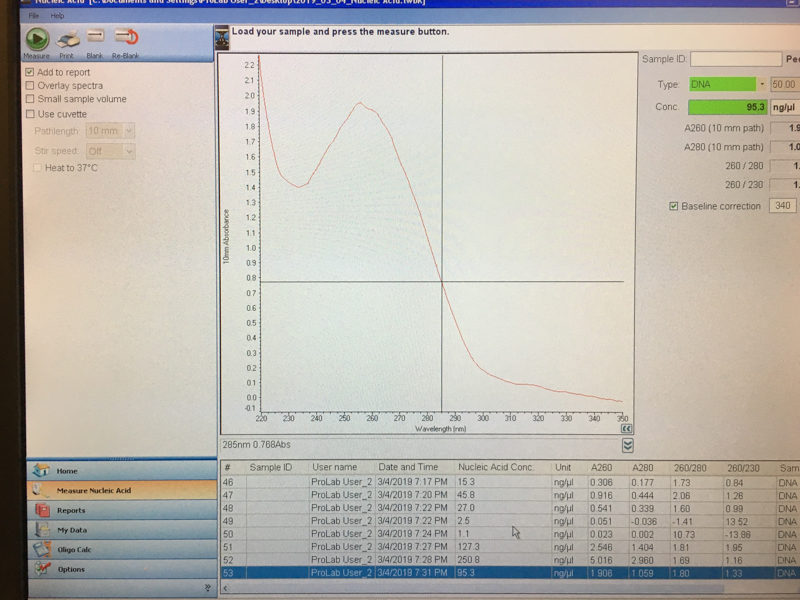
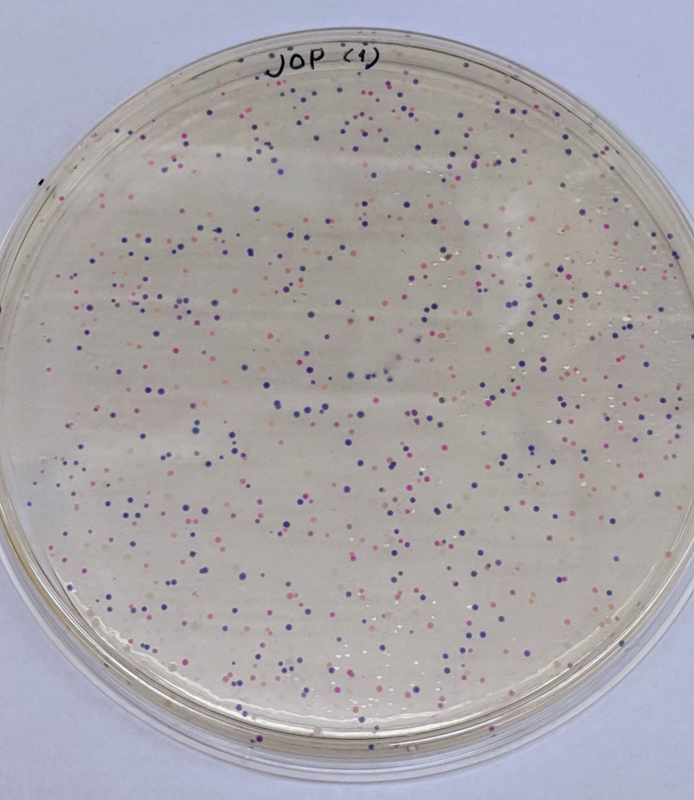
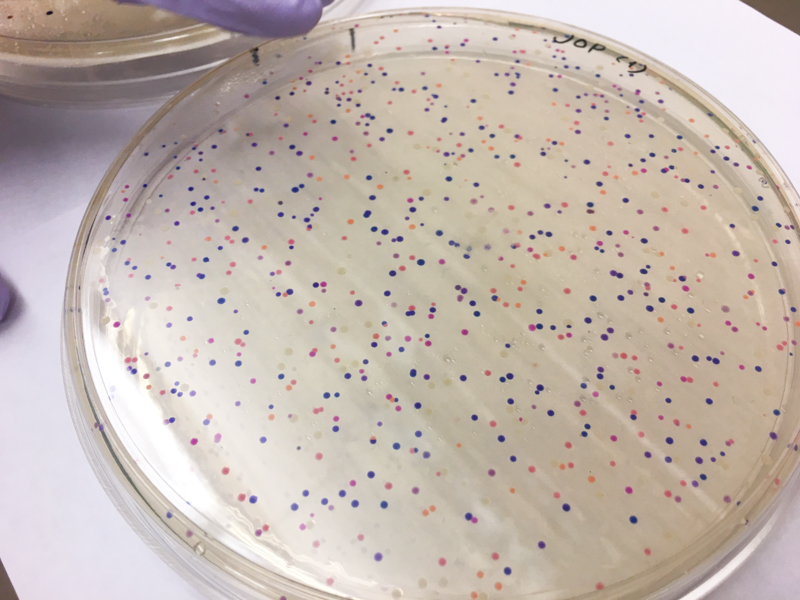
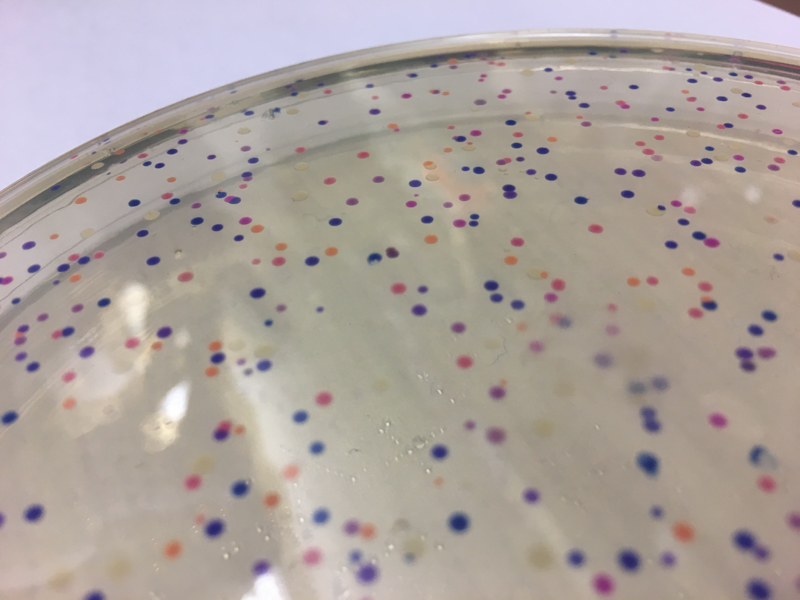
We will be changing the color-generating chromophore of the purple Acropora millepora chromoprotein (amilCP) to a variety of orange, pink, and blue mutants. These divergently-colored genetic variants of amilCP were described by Liljeruhm et al. in 2018. Their strategy to identify where to mutate amilCP was inferred by sequence similarities to the chromophore region that allows for spectral engineering of the structurally-characterized and well-known green fluorescent protein (GFP), which is native to the jellyfish Aequorea victoria (From the experiment protocol).
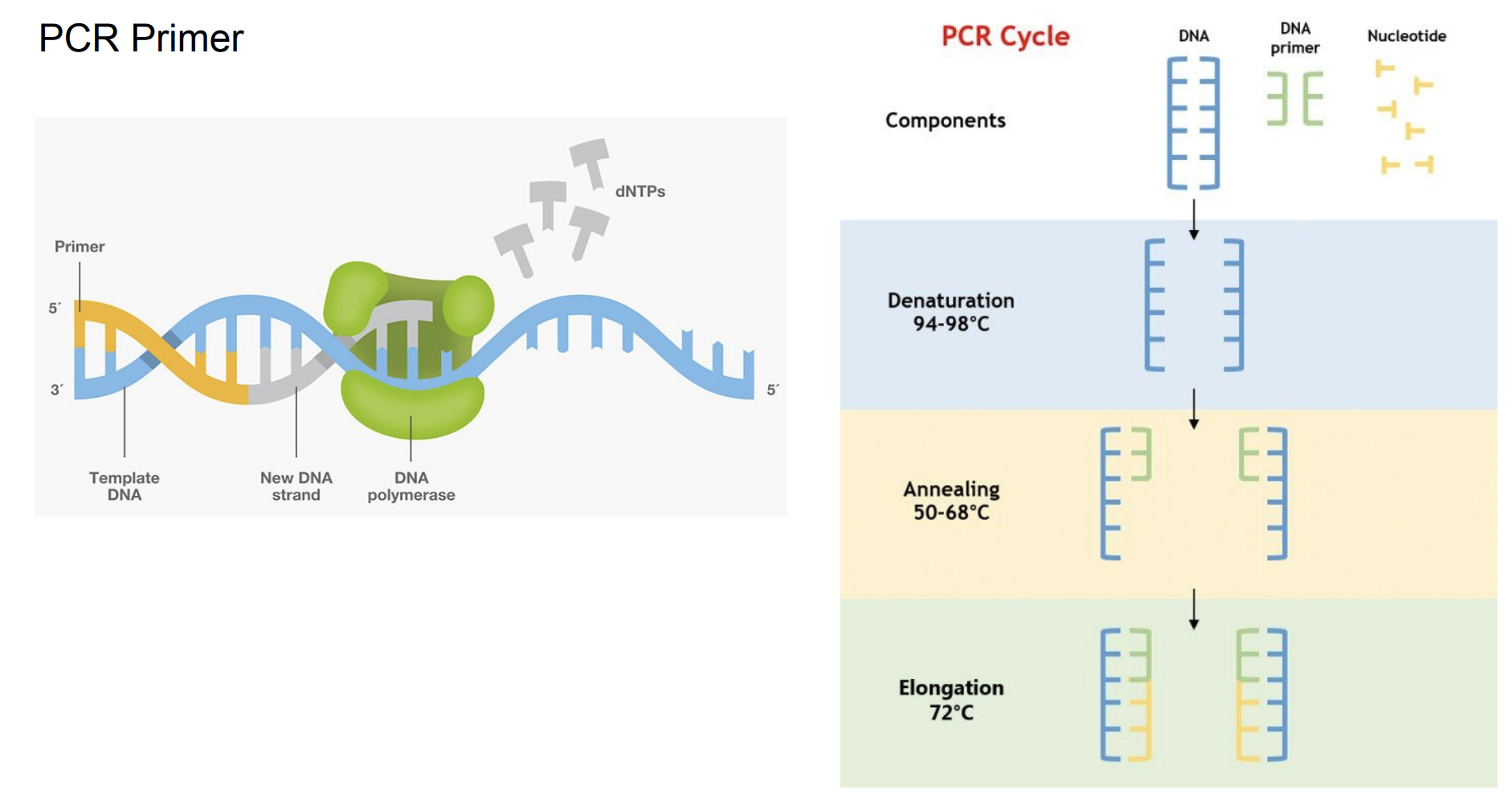
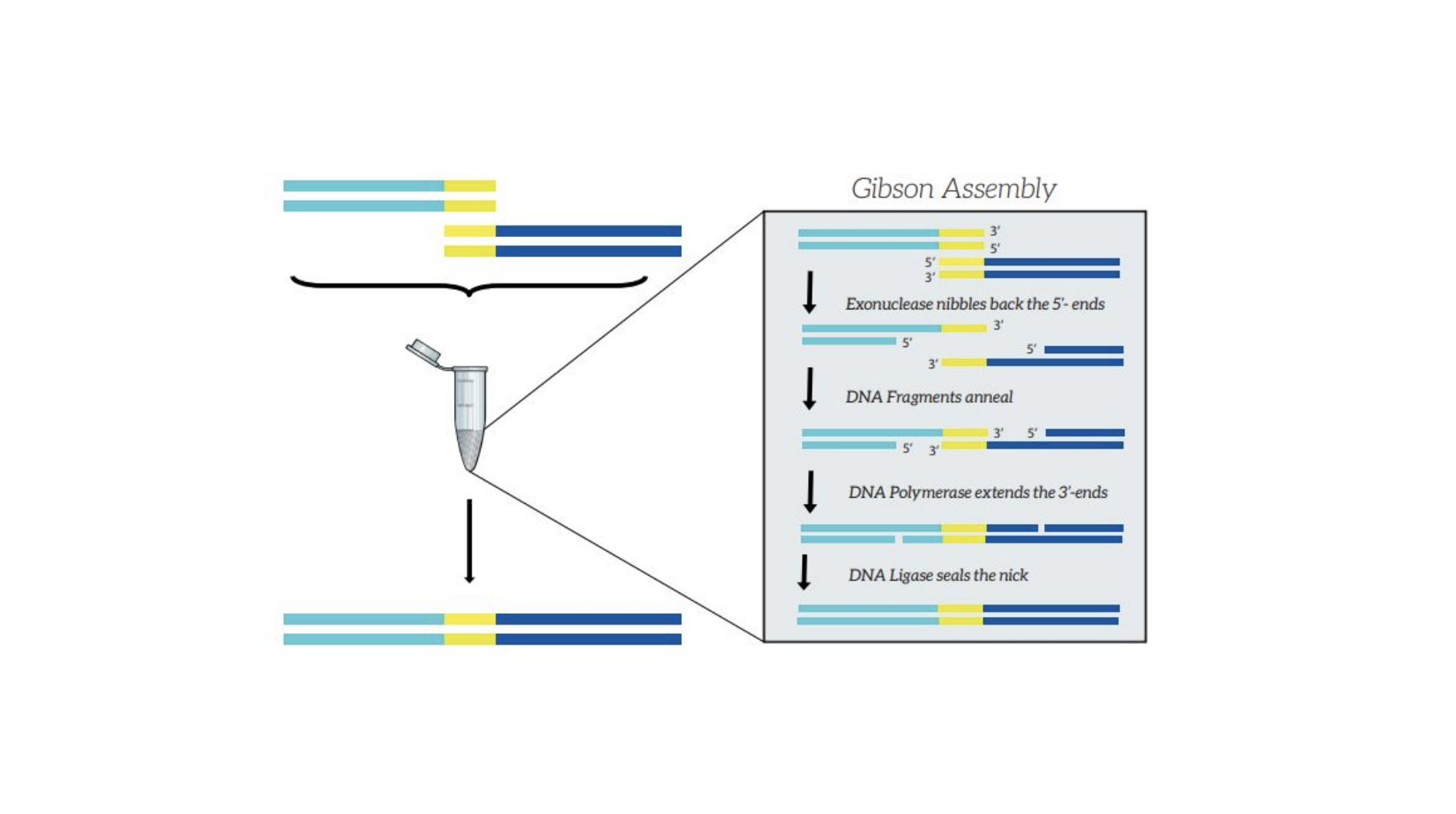
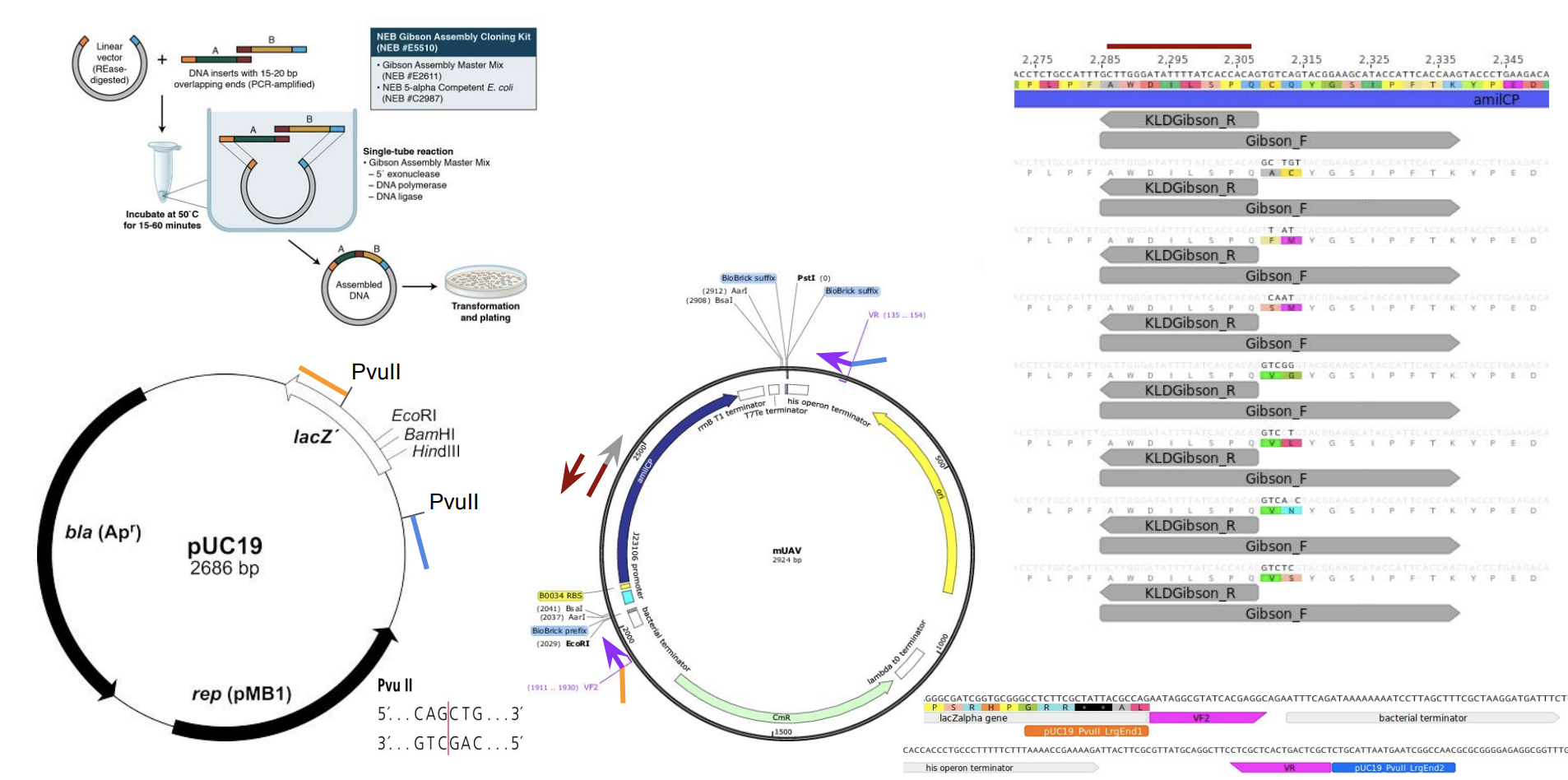
First we prepared the Gibson assembly by using polymerase chain reaction (PCR) to produce two sets of amplicons as inserts and a restriction digest of the common cloning plasmid pUC19 to produce a new backbone (i.e. origin of replication and drug resistance gene). As template, both reactions use the amilCP-encoding plasmid that was miniprepped from the Addgene mUAV sample. One set of amplicons copy the region of the amilCP gene that precedes the chromophore, including the transcription promoter and translation ribosome-binding site (RBS). Another set amplicons copy the region that spans 24 basepairs before the chromophore to just beyond the gene's transcription terminators. The latter includes a diversified chromophore-coding segment dictated by mismatches in the PCR primers with respect to the mUAV DNA template. The amplicon sets both include one end that overlaps by 20-22 bases with distinct ends of the large backbone fragment from the pUC19 digest. Lastly, we will express our colorful variety of amilCP mutants in chemically competent E coli cell (From the experiment protocol).