WEEK 02: PRODUCE DNA LADDERS, in the topic of Bio Design
This journal documents demonstrates the concepts, terms, methodology and workflow of producing DNA ladders using 2 restriction enzymes, the EcoRV and the PstI with Addgene plasmids pPSU1 and pPSU2, as demonstrated by Henrici et al.
Henrici,Ryan C., Turner J. Pecen, James L. Johnston, and Song Tan. 2017. "The PpsuPlasmids For Generating DNA Molecular Weight Markers". Scientific Reports 7 (1).
I. Preparation for Producing DNA ladders
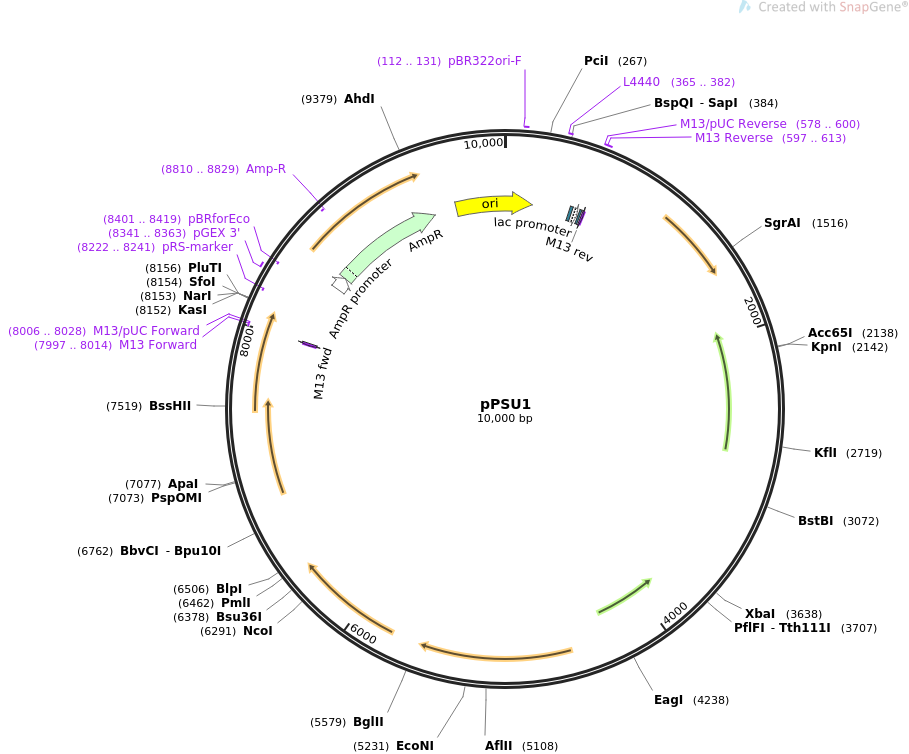
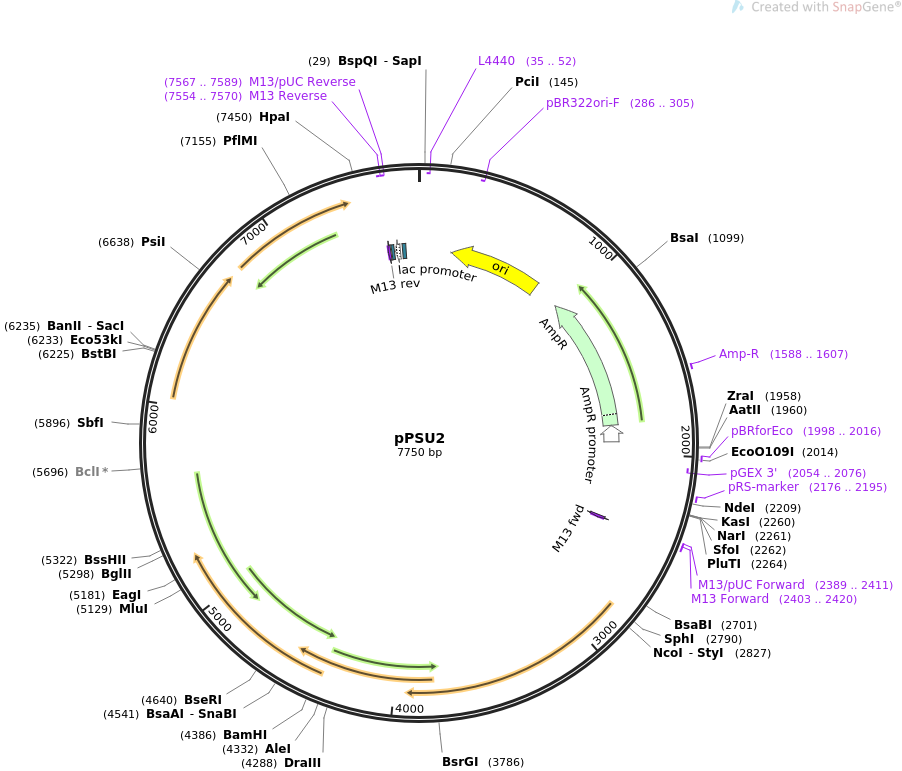
Read and annotate the sequence files for DNA molecules
pPSU1 Sequencing Result:
PstI (7 cut) - 2089, (d=500), 2589, (d=2000), 4589, (d=1000), 5589, (d=1000), 6289, (d=1000), 7089, (d=900), 7989
EcoRV (5 cut) - 634, (d=500), 1134, (d=1000), 2134, (d=1000), 3634, (d=2000), 5634
pPSU2 Sequencing Result:
PstI (7 cut) - 2089, (d=500), 2589, (d=2000), 4589, (d=1000), 5589, (d=1000), 6289, (d=1200), 7089, (d=900), 7989
EcoRV (5 cut) - 634, (d=500), 1134, (d=1000), 2134, (d=1000), 3634, (d=2000), 5634
During the process, fragments of the gene from restriction enzyme EcoRV and PstI will be encoded into the prepared plasmids (bacterial DNA that outside nucleus). We can expect hundreds of copies per cell from these plasmids, much higher than pBR322 (20-30 copies).
II. Harvest pPSU1 and pPSU2 DNA from E. coli culture using the ZymoPure Plasmid Miniprep Kit
We took out pPSU1 and pPSU2 sample from the freezer and added 250 µl of ZymoPURE™ P1 (Red), vortexed.

We then added 250 µl of ZymoPURE™ P2 (Green) and gently inverted the tubes 6-8 times.

After 3min we added 250 µl of ice-cold ZymoPURE™ P3 (Yellow), inverted additional 2-3 times, and incubated the neutralized lysate on ice for 5 minutes.


Then we added 275 µl of ZymoPURE™ binding buffer(balancing PH), filtered fluid, and centrifuged to get our gene (the separated liquid).
After that we got the breakdown mixture while supposed to wash the genetic material of any other debris with ZymoPURE™ Wash I once and ZymoPURE™ Wash II twice; however we accidentally picked up Wash II first, which result in only small yield(10-80ng/ul) on nano spectrophotometer. Thus, we prepared a new gene the next day.
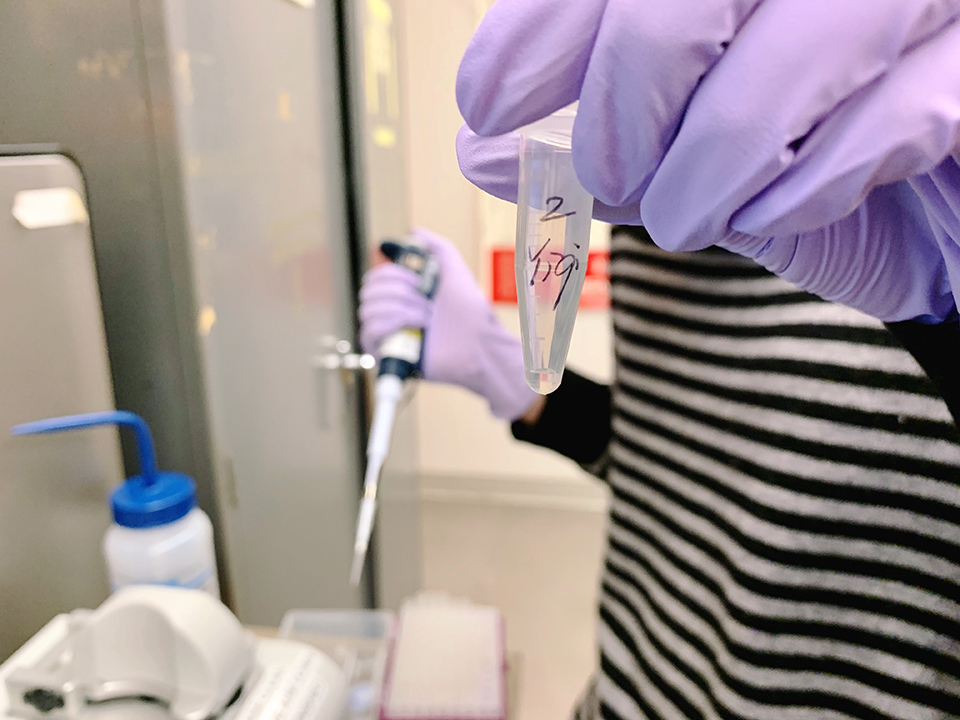
After we prepared our gene, we transferred the sample into a 1.5 ml tube and added 25 µl of DNase/RNase-Free water, incubated, centrifuged to get it ready for nano spectrophotometer.
III. Measure the concentration of pPSU1 and pPSU2 DNA preps using the Nanodrop spectrophotometer

Firstly, we cleaned, wiped dry and blanked (2 ul DI water) the Nanodrop.

Secondly, we run the Nanodrop for our DNA preps.
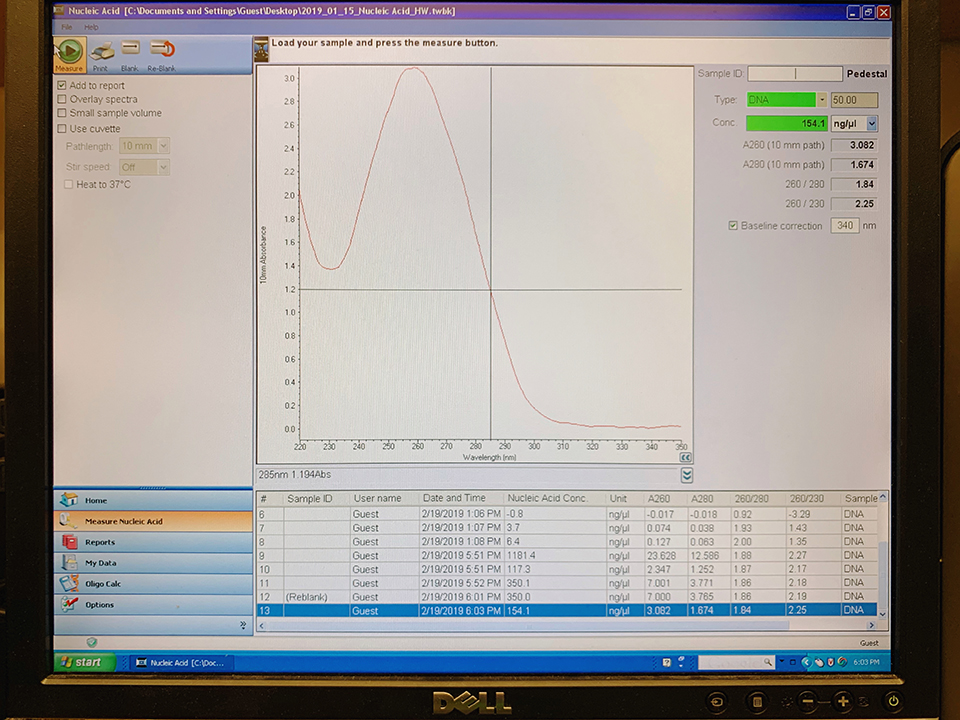
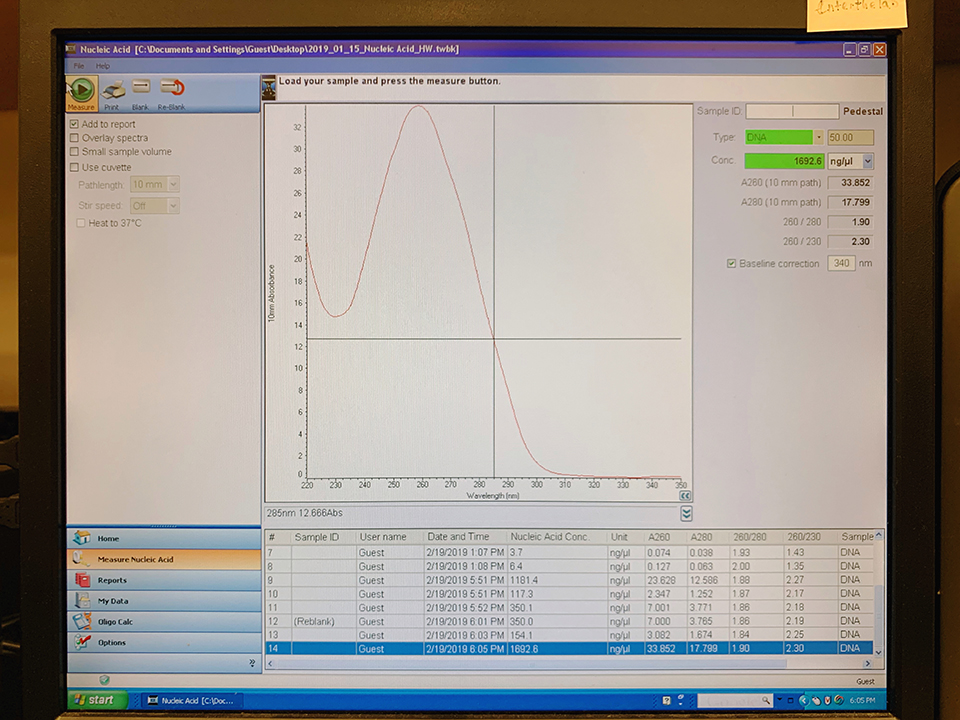
Results from Nanodrop shows the pPSU1 at a yield of 154.1 ng/ul and the pPSU2 at a yield of 1,692.6 ng/ul. Because our pPSU2 yields are higher than 500 ng/ul in concentration, we prepared a dilution prep(2.5ul p2/40ul) with water to 100 ng/ul.
IV. Prepare reaction with EcoRV-HF and PstI-HF
We calculated and mixed our preps as following:
EcoRV-HF (50 ul total)
1 ug pPSU1 DNA = 6.5 ul
1 ug pPSU2 DNA = 10 ul
DNase/RNase-Free Water = 26.5 ul
NEB Buffer CutSmart = 5 ul
NEB EcoRV-HF = 2 ul
PstI-HF (50 ul total)
1 ug pPSU1 DNA = 6.5 ul
1 ug pPSU2 DNA = 10 ul
DNase/RNase-Free Water = 26.5 ul
NEB Buffer CutSmart = 5 ul
NEB PstI-HF = 2 ul
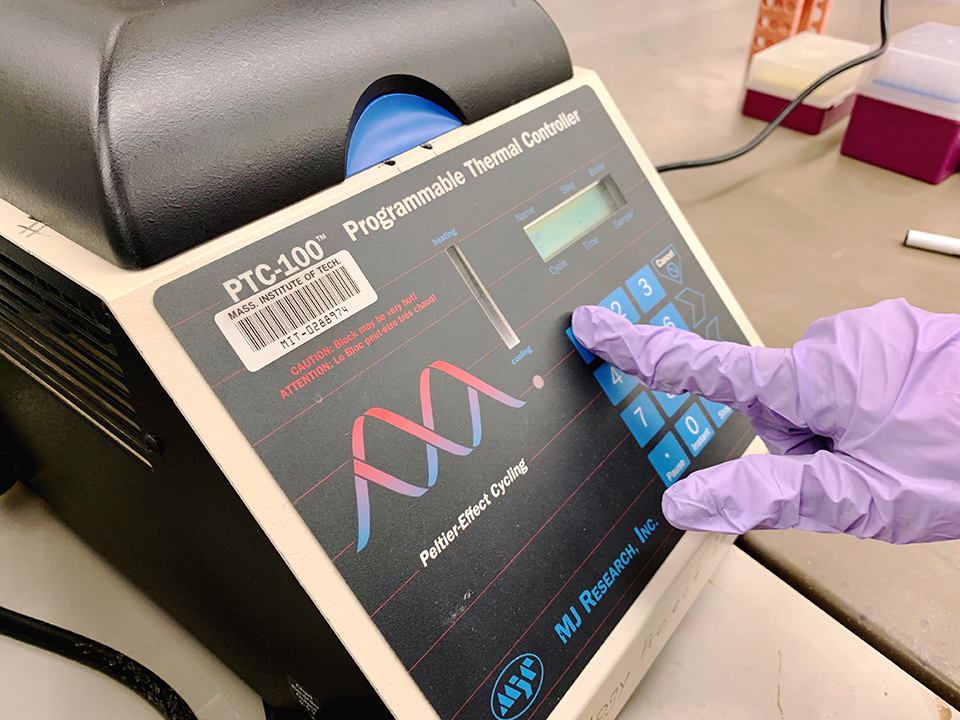
We then incubated preps at 37C for 15 minutes in a thermocycler.
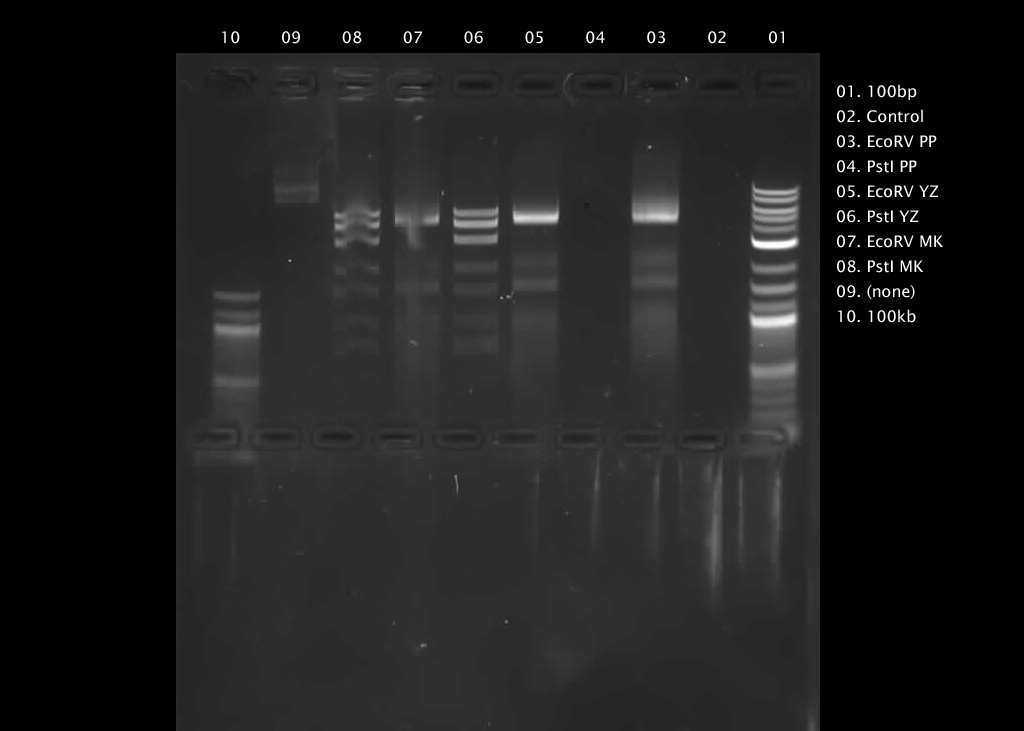
V. Run the reactions alongside 100bp and 1kbp DNA ladder and uncut plasmid controls

We prepared the 1% agarose gel with 1xTAE and 10000xSYBR-Safe DNA stain, poured into rotated gel box and waited 45-60 minutes for gel to set. Then we turned the tray, Filled the box with 1xTAE until the gel is submerged and set up electronics.

In the end, we loaded our preps along with 10 ul 6xGel Loading Dye per lane and ran them at 120V for 1 hour.

Our final gel electrophoresis illustrated the cut locations.