Week 5: Bio-Production
This week we had a homework assignment set by Patrick Boyle from Gingko Bioworks, which was about engineering e. coli so that it produces lycopene, the red pigment found in tomatos. Specifically, by transferring a 3-enzyme pathway, e. coli can be used to turn farnesyl diphosphate to lycopene. The computational tools and databases presented today can also be used to enhance lycopene production in E. coli or even produce different colors, such as the beta-carotene pigment that makes carrots orange.
For our assignment, we used teh TOP10 E. Coli strain from addgene with the pAC-LYC, pAC-LYCipi and pAC-BETAipi plasmids, which contain the genes which enable the metabolic pathways for lycopene productions in the e.coli.
Addgene: pAC-LYC contains 3 genes (crtE, crtI, and crtB) of the carotenoid pathway gene cluster of Erwinia herbicola (Pantoea agglomerans) Eho10 and thereby produces lycopene in Escherichia coli
The Arabidopsis thaliana idi gene is added to the pAC-LYCipi plasmid to boost lycopene production.
The pAC-BETAipi plasmid produces beta-carotene through the addition of the Erwinia herbicola crtY gene.
We have several variables: cultivating the e.coli with the three plasmids, in different growth media (LB, LB + Fructose, 2YT, 2YT + Fructose), across 12 samples.

First we do a lot of pipetting, combining 200ul of each e.coli sample with 5ml of the growth media.
We make two of each combination.

They go into the warm rooms, and are spun for 24 hours to grow the cultures. The grown cultures are transferred into a 96 well plate for the Biotek spectrophotometer.
Each sample is transferred into centrifuge tubes and spun at 13000 for a minute to create a pellet, so that the supernatant can be removed.

Pellets are then mixed with acetone, which dissolves the lycopene in the e. coli.
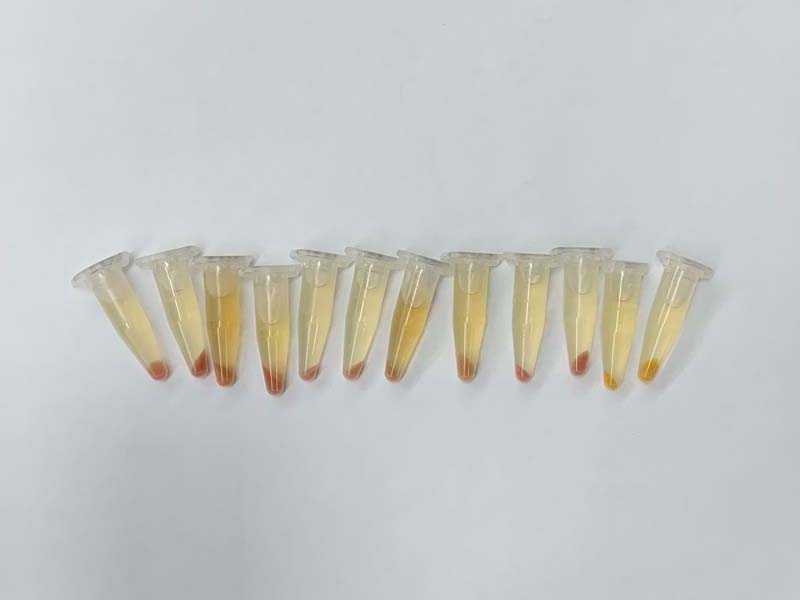
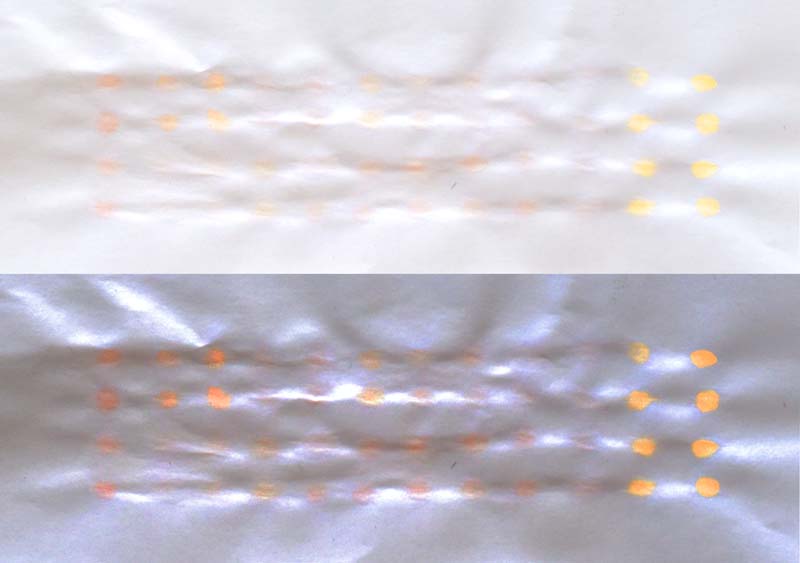
Here we can see how each lycopene sample looks like on normal printer
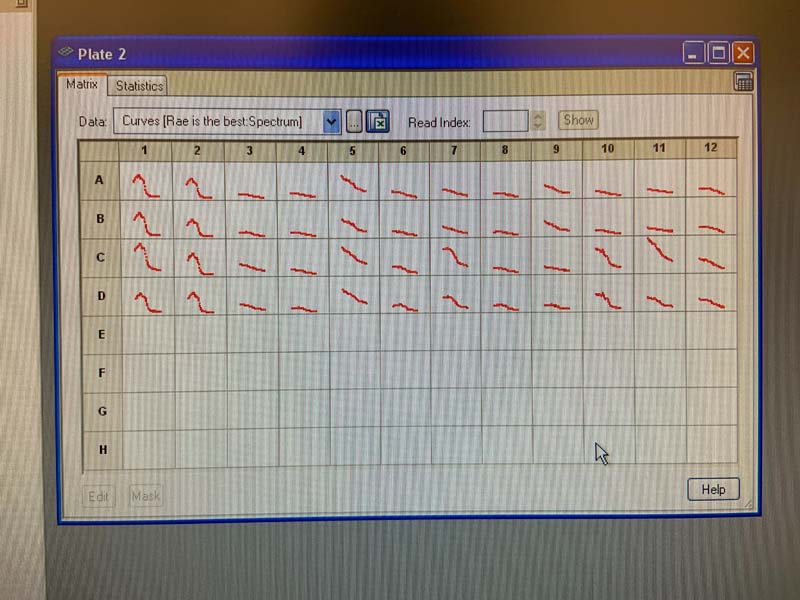
And here we see the spectrophotometry results, in a single graph.
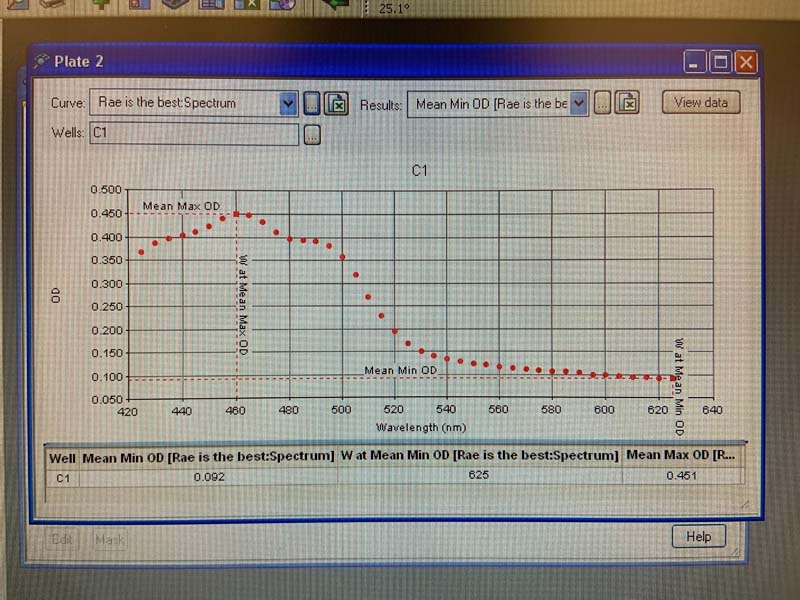
And for the whole plate.