Week 6: Protein Design
How many molecules of amino acids do you take with a piece of 500 grams of meat? (on average an amino acid is ~100 Daltons)
500g meat (beef)
130g protein
~21 amino acids per protein
0.13/(1.6x10^-25) = 8.125x10^23
Protein notes
Amino acids have a carboxylic group and an amino group.
Every amino acid has a carbon, the alpha carbon.
A carboxylic acid is R-COOH, where R is the rest of the molecule (C(=O)OH)
When the amino group is bonded directly to the alpha carbon, its an "alpha amino acid"
Every amino acid has teh Alpha Carbon (Ca), bonded to carboxylic acid, (-COOH); the "amino" is the -NH2 Group; and a hydrogen atom, and the R group.
Ca is a chiral center, meaning this carbon atom is attached to four different groups. All amino acids except glycine, which has no R, is chiral. In glycine R =H.
L and D amino acids are mirrors of each other. Left and right handed. (enantiomers) are those molecules which overlap when two mirrored amino acids are superimposed on one another.
Only L-amino acids make up proteins. Our bodies make them and then they get incorpoated into proteins.
There are 20 common amino acids. they are organized in terms of their R group.
Hydrophobic
Valine
Leucine
Alanine
Isoleucine
Methionine
Proline
Tryptophan
hydrophilic
Serine
Threonine
Cysteine
Glutamine
Aspartic acid
Histidine
Argenine
Amino acids are joined by peptide bonds.
1) a nucleophilic amino group attacks an electrophilic carbonyl group
2) the carbonyl bond reforms with the elimination of a hydroxide ion (H2N becomes N-HH)
2) hydroxide ion abstracts a proton (eliminates water) and the positive charge on nitrogen is neutralized.
Why are there only 20 natural amino acids?
The first amino acids may have started to anchor membranes to RNA structures.
They may have emerged from lightning with water, methane, ammonia and hydrogen.
Meteorites have also be found with many amino acids (86 on Murichison meteorite).
The proteinogenic amino acids, however, were chosen in the RNA world. Amino acids may have been a product of RNA metabolism, and since RNA molecules were the first self-replicators, this would have hugely increased the presence of proteinogenic amino acids in the environment.
The last universal common ancestor was using the same 20 amino acids.
These amino acids have properties to solve particular "jigsaw puzzles", combinations of hydrophobic/hydrophilic with different breanching patterns to make particular surfaces and gaps.
These amino acids also exhibit a wide number of properties when distributed across a space of chemical properties (charge, size and hydrophobicity), ie. they cover all the bases relatively evenly. (Freeland)
Oxygen entering into the equation also made at least 6 of the proteins possible. Some amino acids are much more prone to oxidative degradation. More redox active amino acids could protect cells, "maintaining a libid bilayer integrity in the presence of rising oxygen concentration or in the presence of chemical influences which tend to attack or degrade unsaturated fatty acids"
There are also two other amino acids used in organisms, though not in normal protein synthesis: selenocysteine, and pyrrolsine.
Protein synthesis is carried out by tranlsation, by the cell's ribosome (a very large complex of RNA and protein molecules). Each amino acid is carried by a bespoke transfer RNA (tRNA) molecule, attachd through a hydroxyl group to form an ester. The correct amino acid sequence is translated from messenger RNA molecules through Watson Crick base-pairing with the tRNA molecules. Each tRNA contains a sequence of 3 bases specific to one of hte amino acids. A codon.
The point where nature was unable to create new unique tRNAs that would not be mistaken for others seems to have been at 20 amino acids. In modern biology this allows most amino acids to be coded by more than one codon – the redundancy helping more accurate translation.
The dihedral angle is the angle between every two bonds on the backbone. Every three bonds is a phy, psi and omega dihedral angle.
Side chains come off the alpha carbon. Mostly they are large enough to further restrict parts of the dihedral angles from clashing with carbonyl groups. The presence of side chains makes a big difference to right v left. On left handed helices the sidechain bond -R and the carbonyl bond -O is much closer, creating much greater steric hinderance.
https://www.quora.com/Why-are-most-alpha-helices-in-proteins-right-handed
Where did amino acids come from before enzymes that make them, and before life started?
Mixing water, ammonia, methane and hydrogen with lightning can produce amino acids. some may have come from space (glycine!)
What do digital databases and nucleosomes have in common?
A nucleosome is a basic unit of DNA packaging in eukaryotes (cells with a nucleus in a membrane), consisting of DNA segment wrapped around a sequence of 8 histone protein cores.
The histones order DNA into structural units (ribosomes). Basically they keep DNA in an order, and form the repeating units (beads in the string) of chromatin which packs long DNA chains into a dense shape and a smaller volume. They are like cells in a database, or units in a linear array of sorts.
Pick any protein (from any organism) of your interest that has a 3D structure and answer the following questions.
SCALLOP MYOSIN REGULATORY DOMAIN
Briefly describe the protein you selected and why you selected it.
It is the muscle protein of a scallop
Identity the amino acid sequence of your protein.
MNIDFSDPDFQYLAVDRKKLMKEQTAAFDGKKNCWVPDEKEGFASAEIQSSKGDEITVKI VADSSTRTVKKDDIQSMNPPKFEKLEDMANMTYLNEASVLYNLRSRYTSGLIYTYSGLFC IAVNPYRRLPIYTDSVIAKYRGKRKTEIPPHLFSVADNAYQNMVTDRENQSCLITGESGA GKTENTKKVIMYLAKVACAVKKKDEEASDKKEGSLEDQIIQANPVLEAYGNAKTTRNNNS SRFGKFIRIHFGPTGKIAGADIETYLLEKSRVTYQQSAERNYHIFYQICSNAIPELNDVM LVTPDSGLYSFINQGCLTVDNIDDVEEFKLCDEAFDILGFTKEEKQSMFKCTASILHMGE MKFKQRPREEQAESDGTAEAEKVAFLCGINAGDLLKALLKPKVKVGTEMVTKGQNMNQVV NSVGALAKSLYDRMFNWLVRRVNKTLDTKAKRNYYIGVLDIAGFEIFDFNSFEQLCINYT NERLQQFFNHHMFILEQEEYKKEGIAWEFIDFGMDLQMCIDLIEKPMGILSILEEECMFP KADDKSFQDKLYQNHMGKNRMFTKPGKPTRPNQGPAHFELHHYAGNVPYSITGWLEKNKD PINENVVALLGASKEPLVAELFKAPEEPAGGGKKKKGKSSAFQTISAVHRESLNKLMKNL YSTHPHFVRCIIPNELKQPGLVDAELVLHQLQCNGVLEGIRICRKGFPSRLIYSEFKQRY SILAPNAIPQGFVDGKTVSEKILAGLQMDPAEYRLGTTKVFFKAGVLGNLEEMRDERLSK IISMFQAHIRGYLIRKAYKKLQDQRIGLSVIQRNIRKWLVLRNWQWWKLYSKVKPLLSIA RQEEEMKEQLKQMDKMKEDLAKTERIKKELEEQNVTLLEQKNDLFLQLQTLEDSMGDQEE RVEKLIMQKADFESQIKELEERLLDEEDAAADLEGIKKKMEADNANLKKDIGDLENTLQK AEQDKAHKDNQISTLQGEISQQDEHIGKLNKEKKALEEANKKTSDSLQAEEDKCNHLNKL KAKLEQALDELEDNLEREKKVRGDVEKAKRKVEQDLKSTQENVEDLERVKRELEENVRRK EAEISSLNSKLEDEQNLVSQLQRKIKELQARIEELEEELEAERNARAKVEKQRAELNREL EELGERLDEAGGATSAQIELNKKREAELLKIRRDLEEASLQHEAQISALRKKHQDAANEM ADQVDQLQKVKSKLEKDKKDLKREMDDLESQMTHNMKNKGCSEKVMKQFESQMSDLNARL EDSQRSINELQSQKSRLQAENSDLTRQLEDAEHRVSVLSKEKSQLSSQLEDARRSLEEET RARSKLQNEVRNMHADMDAIREQLEEEQESKSDVQRQLSKANNEIQQWRSKFESEGANRT EELEDQKRKLLGKLSEAEQTTEAANAKCSALEKAKSRLQQELEDMSIEVDRANASVNQME KKQRAFDKTTAEWQAKVNSLQSELENSQKESRGYSAELYRIKASIEEYQDSIGALRRENK NLADEIHDLTDQLSEGGRSTHELDKARRRLEMEKEELQAALEEAEGALEQEEAKVMRAQL EIATVRNEIDKRIQEKEEEFDNTRRNHQRALESMQASLEAEAKGKADAMRIKKKLEQDIN ELEVALDASNRGKAEMEKTVKRYQQQIREMQTSIEEEQRQRDEARESYNMAERRCTLMSG EVEELRAALEQAERARKASDNELADANDRVNELTSQVSSVQGQKRKLEGDINAMQTDLDE MHGELKGADERCKKAMADAARLADELRAEQDHSNQVEKVRKNLESQVKEFQIRLDEAEAS SLKGGKKMIQKLESRVHELEAELDNEQRRHAETQKNMRKADRRLKELAFQADEDRKNQER LQELIDKLNAKIKTFKRQVEEAEEIAAINLAKYRKAQHELEEAEERADTADSTLQKFRAK SRSSVSVQRSSVSVSASN
How long is it? What is the most frequent amino acid?
1,938
How many protein sequence homologs are there for your protein?
20
https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=7273
There is a myosin superfamily of muscle fibres
http://www.rcsb.org/structure/1WDC
It is solved and a good quality structure.
Open the structure of your protein in any 3D molecule visualization software
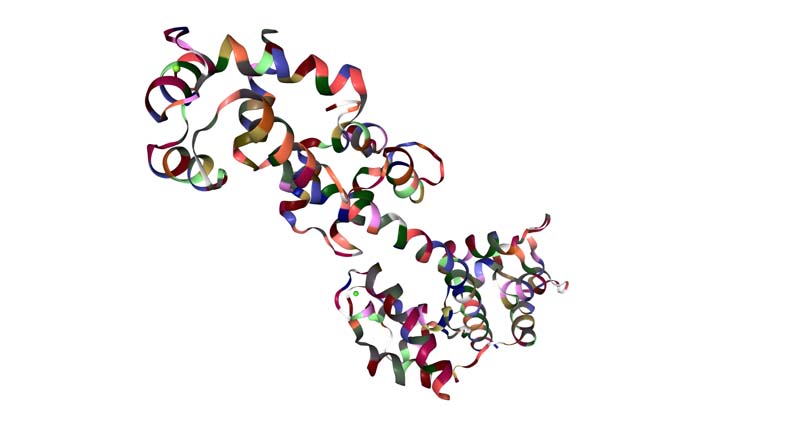
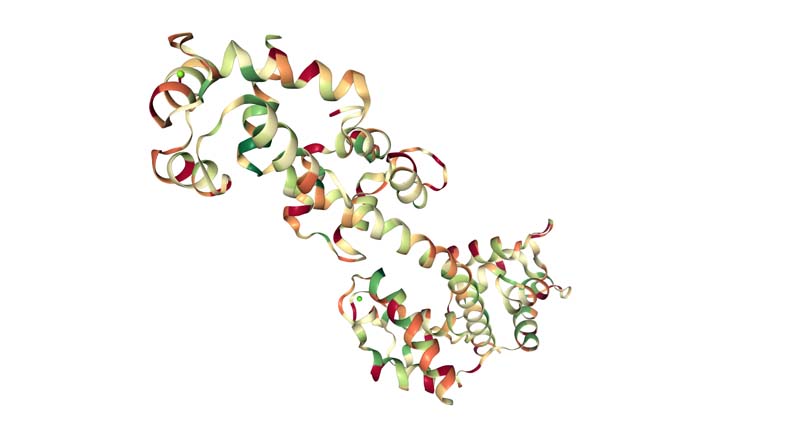
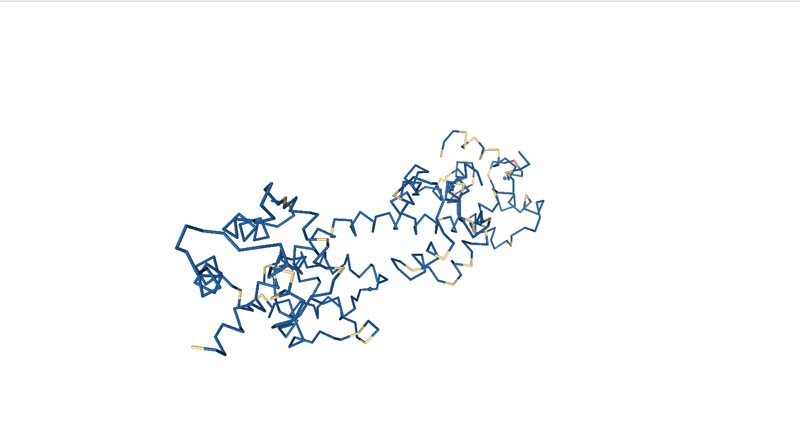
Visualize the protein as "cartoon", "ribbon" and "ball and stick".
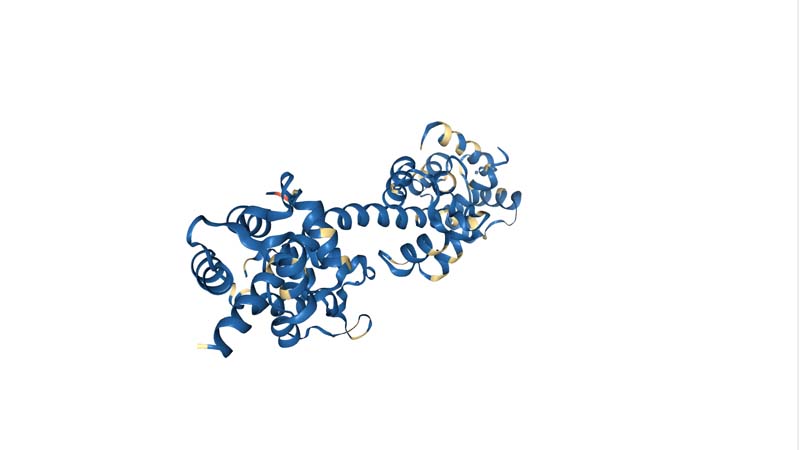
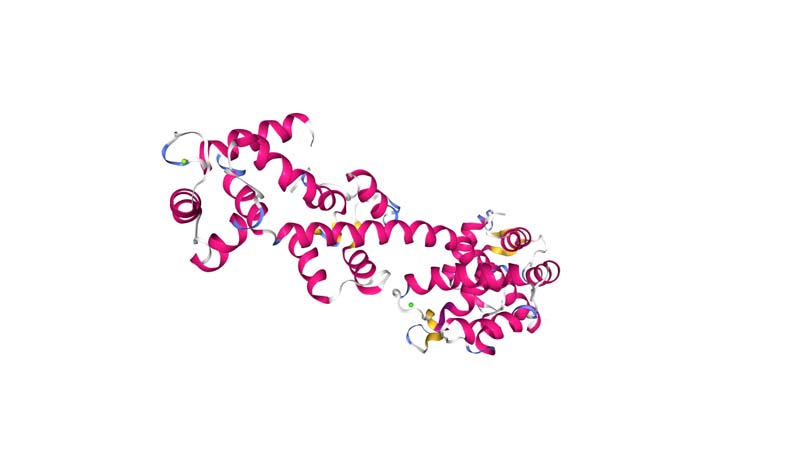
Color the protein by secondary structure. Does it have more helices or sheets?
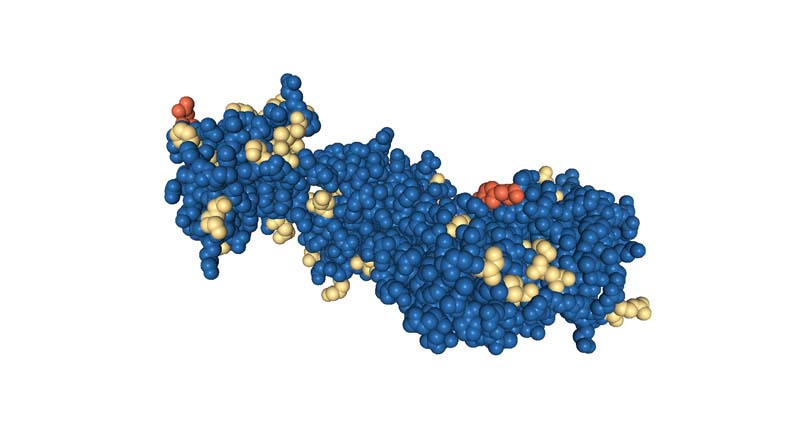
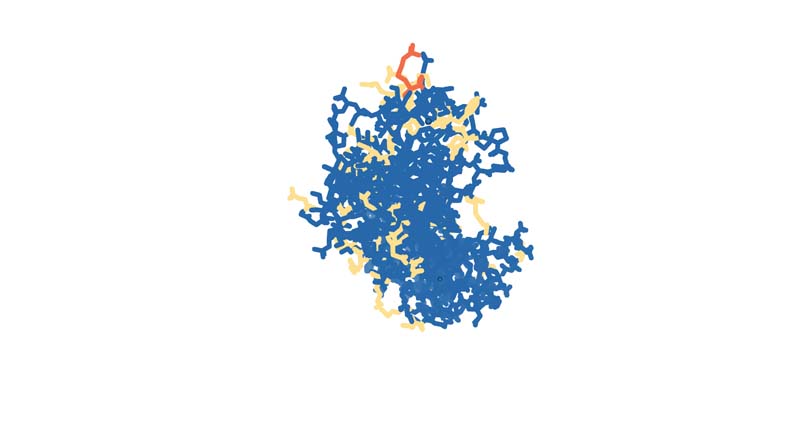
Color the protein by residue type. What can you tell about the distribution of hydrophobic vs hydrophilic residues?
Visualize the surface of the protein. Does it have any "holes" (aka binding pockets)?