Protein Design
by Shuguang Zhang and Thras Karydis
Part A: Protein analysis
Question: How many molecules of amino acids do you take with a piece of 500 grams of meat? (on average an amino acid is ~100 Daltons)
Answer:
Assuming that meat is 26% proteins,
then 500g of meat contains 130g of protein
which equates to 7.829e+25 Daltons
and if each Amino Acid is 100 Daltons then
That is 7.829e+23 individual Amino Acids in 500g meat
That's a hard number to visualize,
So I calculated that is about
1.5 Quintillion amino acid molecules in a mm3 of meat (which still doesnt help)
Question: Why are there only 20 natural amino acids?
Answer: There are many theories but none are especially strong; it
appears to me that we really don't know. The simplist idea is that they just happened
to be the set which were in use by the last common ancestor millions of
years ago, and so have just remained in all life. Regardless it appears that 20
is a stable number which has neither increased or decreased any time that we know of.
DNA comes in four types of nucleotides (A,T,G and C) which makes 64 possible triplet
combinations. Therefore, life would have the potential to make more than our simple 20
amino acids. This would suggest that other Amino Acids are not necessary (perhaps they
existed once but just went out of fashion to save energy) and that 20 turns out to be
needed to create the current mix of variety which we are used to on Earth.
Question: Why most molecular helices are right handed?
Answer: It appears that the reasons are not known with certainty but most explainations are very complicated and related to the molecular stability of the righ handed geometry. A simple explaination would be that the left handed helix results in the amino acid side chains being positioned next to the C=O group, making the structure over-crowded. The right handed version of the helix places side chains next to the much smaller N-H, which is simply a better fit.
Question: Where did amino acids come from before enzymes that make them, and before life started?
Answer: The Miller-Urey experiment of 1952 tried to recreate the chemical conditions on early earth (ammonia, hydrogen, methane, and water vapor plus electrical sparks) and published results showing that from this simple combination of conditions, eleven standard amino acids were formed. In 2007, scientists opened and tested sealed vials preserved from the original experiments and showed that there were actually well over 20 amino acids - far more than has been reported.
Question: What do digital databases and nucleosomes have in common?
Answer: A nucleosome is a packaging unit of DNA. Its formed when eigt histone proteins attach to the string of DNA. The DNA tightly loops around the histone proteins to form a dense package. Neighbouring nucleosomes form clusters and then the clusters coil around and the resulting packed structure is known as chromatin.
A nucleosome is therefore a method of packaging DNA, which is itself just a mechanism
to store genetic data. A neuclesome is similar to a database in that they are both storage
mechanisms for data which is organised in a way that can support read/write functions.
Specifically, a neuclesome is like a sub-unit of data storage within a database -
for example more like a data table.
Pick any protein (from any organism) of your interest that has a 3D structure and answer the following questions.
Question: Briefly describe the protein you selected and why you selected it.Answer: Cows Milk (Bos
Taurus) contains 3.3% total proteins and these proteins are made up of 82% caesin proteins
and 18% of whey proteins:
The casein family of protein consists of several types of casein proteins, each with its
own amino acid composition, genetic variations, and functional properties. The four types of
caesins are: α-s1, α-s2, ß and K. Whereas the 18% which is termed whey actually consists of
several more proteins, which are collectively refered to as whey. The composition of Whey
is β-lactoglobulin: ~ 10%, α-Lactalbumin: ~2%, Serum albumin: ~ 1%, Immunoglobulins: ~ 2%
and Other Proteins: ~ 2%.
I selected to look at Beta Caesin as its the most common of the caesin
proteins in milk, making up about 35% of caesins in cows milk.
Question: Identity the amino acid sequence of your protein.
Answer: >sp|P02666|CASB_BOVIN Beta-casein OS=Bos taurus OX=9913 GN=CSN2 PE=1 SV=2
MKVLILACLVALALARELEELNVPGEIVESLSSSEESITRINKKIEKFQSEEQQQTEDEL
QDKIHPFAQTQSLVYPFPGPIPNSLPQNIPPLTQTPVVVPPFLQPEVMGVSKVKEAMAPK
HKEMPFPKYPVEPFTESQSLTLTDVENLHLPLPLLQSWMHQPHQPLPPTVMFPPQSVLSL
SQSKVLPVPQKAVPYPQRDMPIQAFLLYQEPVLGPVRGPFPIIV
Question: How long is it? What is the most frequent amino acid?
Answer: It is 224 amino acids long. The most frequent amino acid is Proline which occurs 35 times.
Question: How many protein sequence homologs are there for your protein? Hint: Use the pBLAST tool to search for homologs and ClustalOmega to align and visualize them.
Answer: A basic blast protein search when restricte
There were homolg sequences to my beta caesin. This is expected because it is a milk protein, and to the
'Bos Taurus' genus returns 55 homologous entries. However, these are all Bos Taurus and duplicates of the Beta Caesin protein.
There was one protein which was a 7% Query Cover homolog within the Bos Taurus family and it was the 'alpha-S1-casein
isoform X18' protein. Here is the graphical comparison of the two protein sequences visualized with
Clustal Omegal.

For example, Bubalus bubalis (Water buffalo) had a 100% Query Cover match. This is interesting because Cows
(Genus:Bos) and Water Buffalo (Genus: Bubalus) are from the same subfamily (Subfamily: Bovinae) but belong
to different Genus. Clearly their milk is very similar if not identical.
>QHB80269.1 casein beta [Bubalus bubalis]
MKVLILACLVALALARELEELNVPGEIVESLSSSEESITHINKKIEKFQSEEQQQTEDELQDKIHPFAQTQSLVYPFPGPIPNSLPQNIPPLTQTPVVVPPFLQPEIMGVSKVKEAMAPKHKEMPFPKYPVEPFTESQSLTLTDVENLHLPLPLLQSWMHQPPQPLPPTVMFPPQSVLSLSQSKVLPVPQKAVPYPQRDMPIQAFLLYQEPVLGPVRGPFPIIV
>sp|P02666|CASB_BOVIN Beta-casein OS=Bos taurus OX=9913 GN=CSN2 PE=1 SV=2
MKVLILACLVALALARELEELNVPGEIVESLSSSEESITRINKKIEKFQSEEQQQTEDELQDKIHPFAQTQSLVYPFPGPIPNSLPQNIPPLTQTPVVVPPFLQPEVMGVSKVKEAMAPKHKEMPFPKYPVEPFTESQSLTLTDVENLHLPLPLLQSWMHQPHQPLPPTVMFPPQSVLSLSQSKVLPVPQKAVPYPQRDMPIQAFLLYQEPVLGPVRGPFPIIV
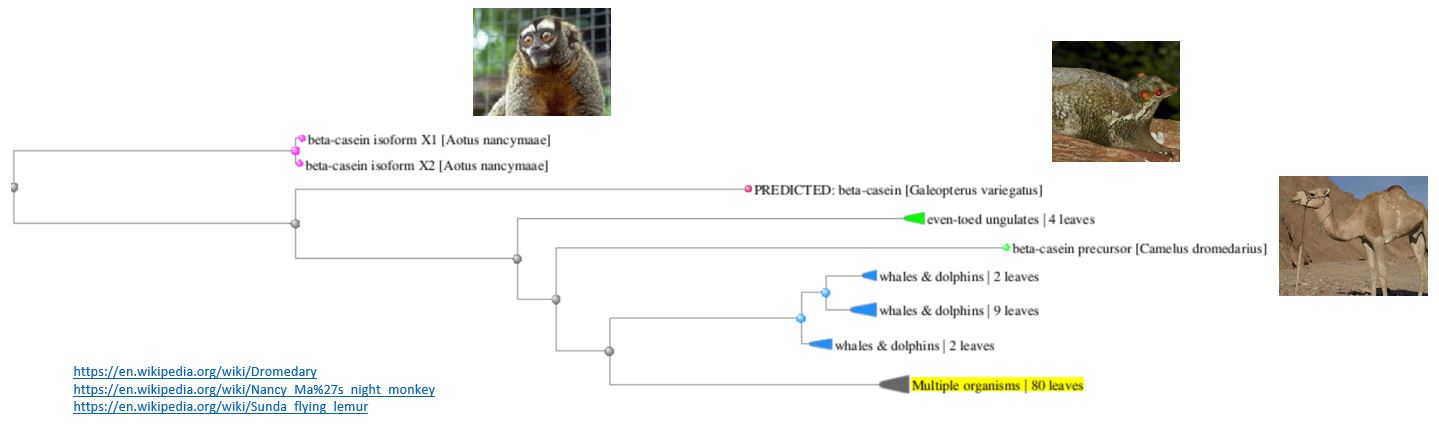
Interestingly, there were many species where there was a high degree of protein homology - I guess
because milk is similar between species. For example, wild boar (Sus scrofa) showed a 99% Query Cover
match (meaning 99% of the amino acids in location are the same).
Here is the Tree View of the homologs of my Beta Caesin protein - you can see that many different mammals
feature.
Question: Does your protein belong to any protein family?
Answer: Yes, it belongs to the Beta Caesin Family with 69 other proteins.
Identify the structure page of your protein in RCSB
I don't think that it is there. But I was able to find it at this website which is a Bovine Milk Proteom Database resource. (reliability unknown)
Question: When was the structure solved? Is it a good quality structure?
Answer: The sequence in UNIPROT says "Last modified:July 1, 1989 - v2" which I assume is the date of solving. I don't know how to tell if its is classed as a 'good quality structure'.
Question: Are there any other molecules in the solved structure apart from protein?
Answer: I don't know
Question: Does your protein belong to any structure classification family?
Answer: There are other types of Beta Caesin proteins - maybe they are a structure classification family? (unsure)
Open the structure of your protein in any 3D molecule visualization software
Question: Visualize the protein as "cartoon", "ribbon" and "ball and stick".
Question: Color the protein by secondary structure. Does it have more helices or sheets?
Answer: It appears that there are no sheets but six helixes.
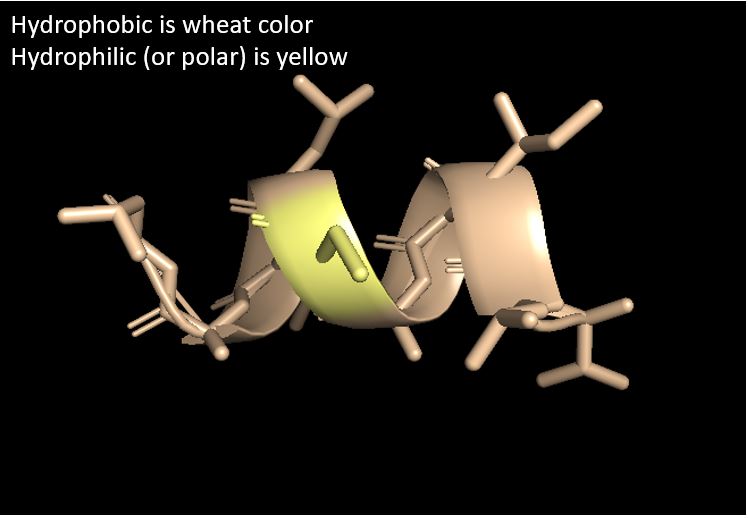
Question: Color the protein by residue type. What can you tell about the distribution of hydrophobic vs hydrophilic residues?
Answer: Most of the structure is hydrophobic with a small part of hydrophilic
in the middle.
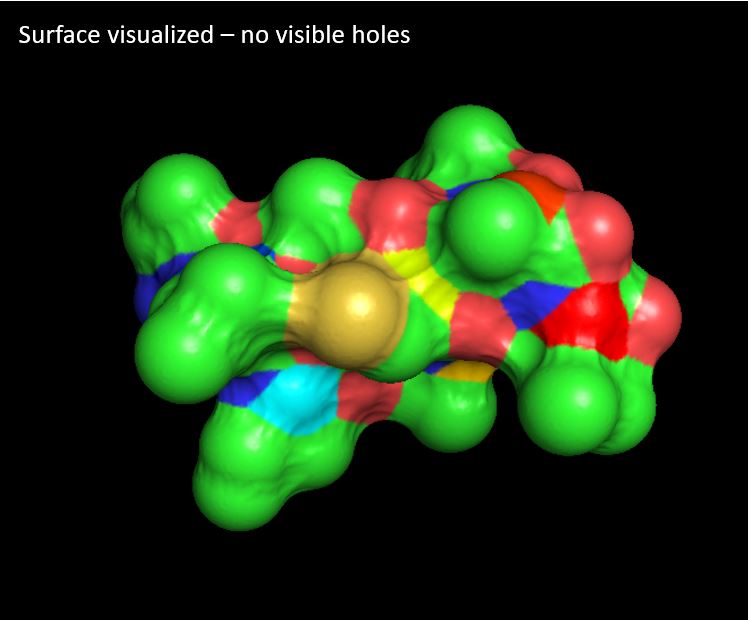
Question: Visualize the surface of the protein. Does it have any "holes" (aka binding pockets)?
Answer: I can't see any holes (aka binding pockets) - low degree of confidence if I have done this question correctly.
Part B: Protein Folding
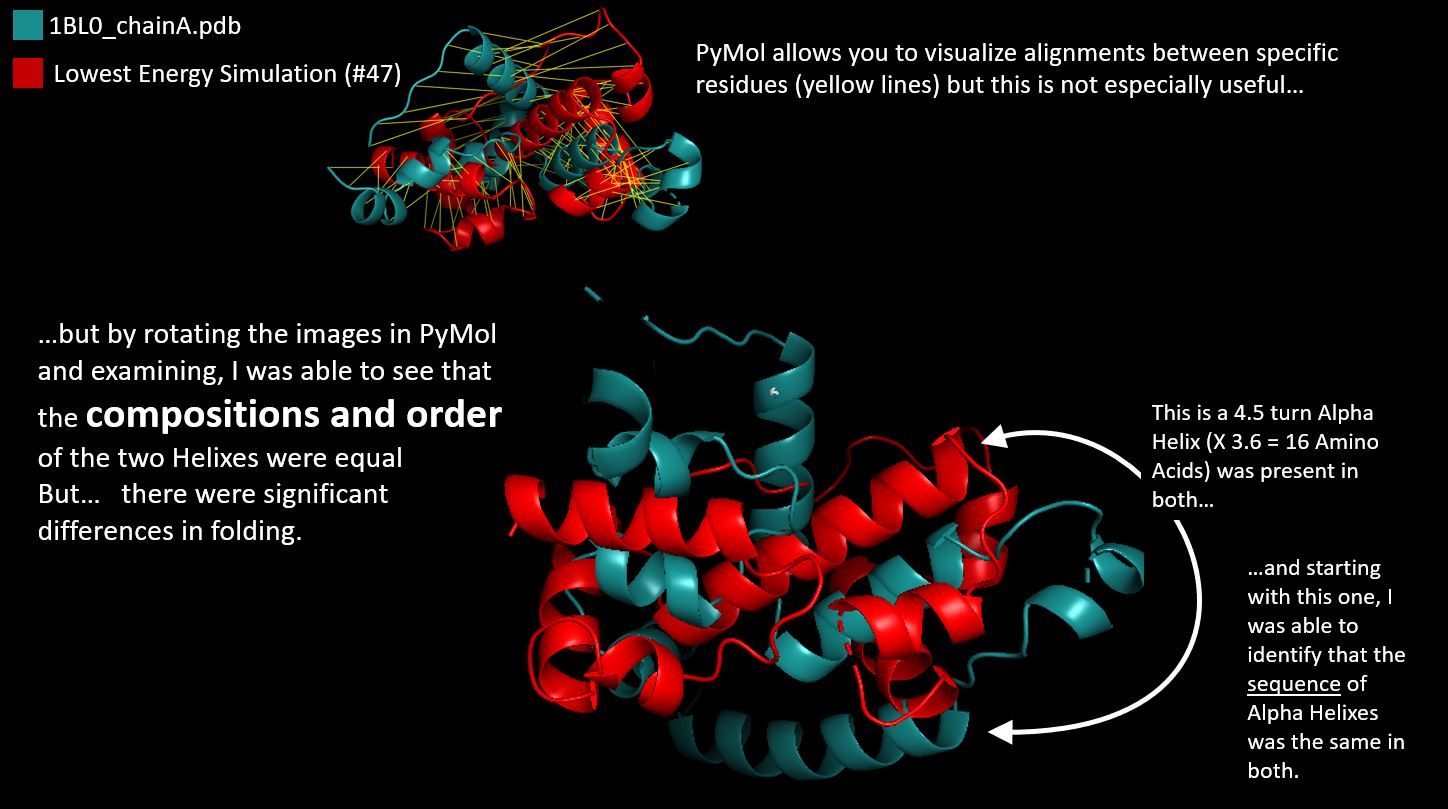
Pyrosetta FileQuestion: Compare the Native Protein with the Lowest Energy Score Simulation
Answer: The lowest Energy Simulation had a score of -261
and I used PyMol to compare with the native. Of course the Alpha Helixes were the same in sequence,
but there were some (small) folding differences which led to a different structure.
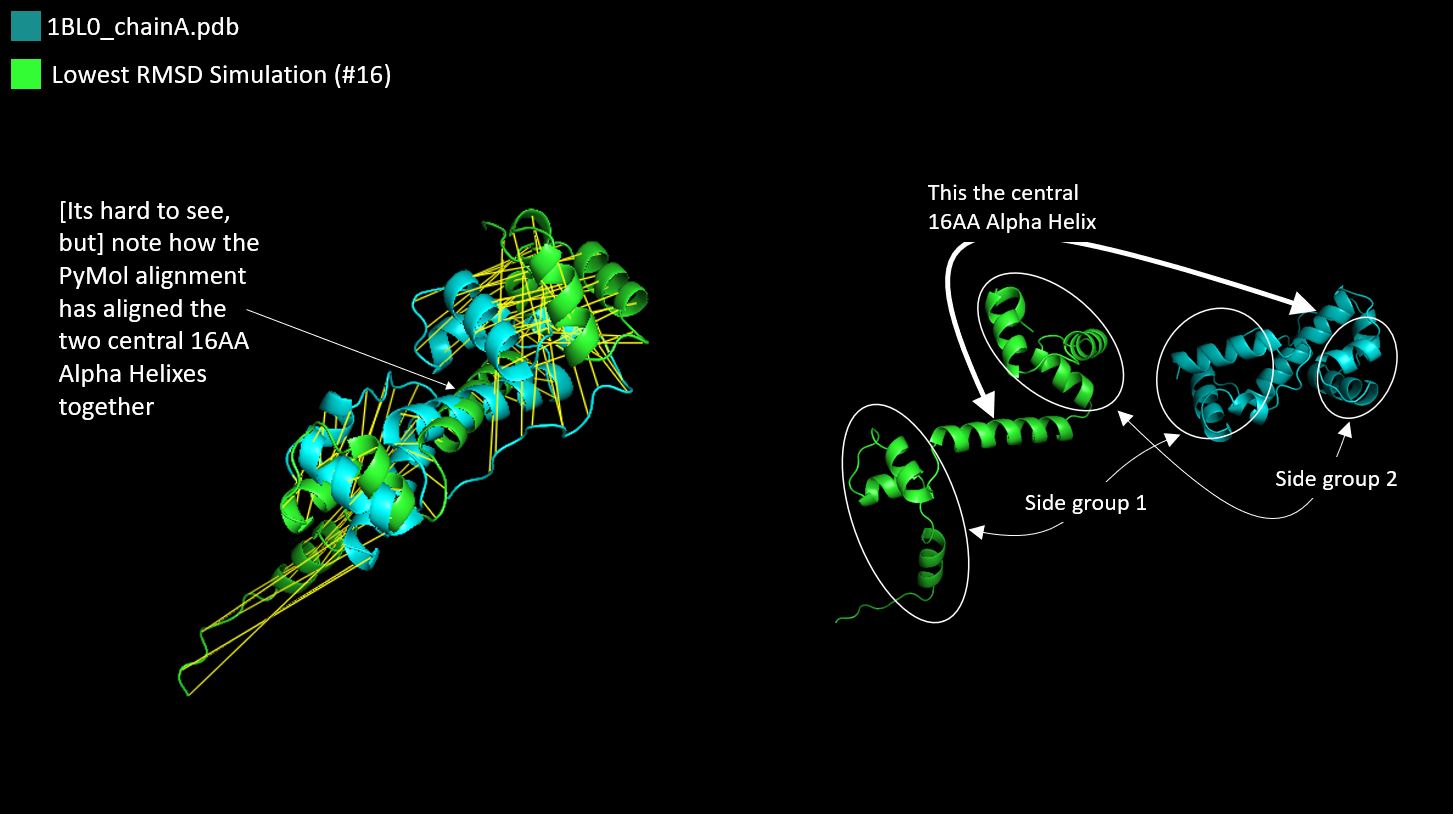
Question: Compare the Native Protein with the Lowest RMSD Simulation
Answer: The lowest RMSD Simulation had a score of 8.943578.
This time the folding differences appeared to be smaller and I felt
that the structures were easier to compare.
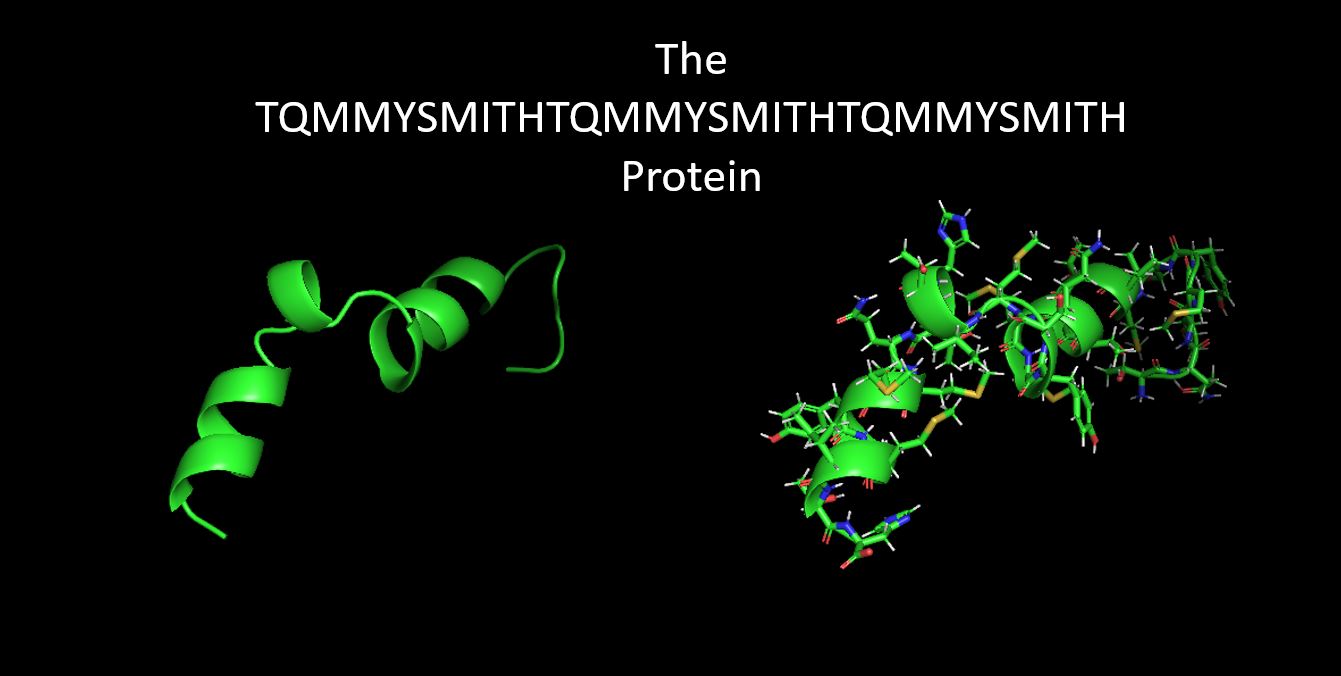
Question: Fold your own sequence!
Answer: I created a new protein with an AA Sequence:
TQMMYSMITHTQMMYSMITHTQMMYSMITH
With the AA Seq repeating 3X: threonine > glutamine > methionine > methionine
> tyrosine > serine > methionine > isoleucine > threonine > histidine
Introducing the West Indian Ocean Coelacanth which has "TQMMYSMIT" in its Protein.

and, my custom designed protein, based on the PyRosette simulation of the specific sequemnce and the resulting
lowest energy score which means that this specific sequence of folds is the most likely to
occur if this protein was really produced.