Next Generation Synthesis
supported by Joe Jacobson, Noah Jakimo, Suryateja Jammalamadaka, Jessica Weber, Parnian Barekatain, Devora Najjar and Berenice Estrada
- Chromophore – Part of a molecule responsible for its color
- Acropora Millipore (Amil CP) - a species of coral
- Chromoprotein – a type of protein which is colored, and responsible for coloration in corals
- Amplicons - a piece of DNA that is the source and/or product of amplification with PCR
- Backbone – in our case, we use this to refer to the part of DNA in plasmid which contains the ORI and the selection marker gene
- Origin of replication - sequence in a genome at which replication is initiated
- Transcription promoter - region of DNA near the transcription start sites of gene, that leads to initiation of transcription
- Ribosome binding site (RBS) - sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of protein translation
This week is all about Jelly Fish, and specifically a protein which exists in a jellyfish which is native to the waters off North America called the Aequorea Victoria (A.K.A. the crystal jelly). The protein is the color-generating chromophore of the Acropora millepora chromoprotein (amilCP) which makes a purple color. Liljeruhm et al in 2018 described how a short 6 base pair sequence is responsible for the color, and by changing this sequence then the protein will express different colors. My aim this week is to change this color generating sequence in order to generate some plate displays of different colors.
See below the Aequorea Victoria and note the purple glow.

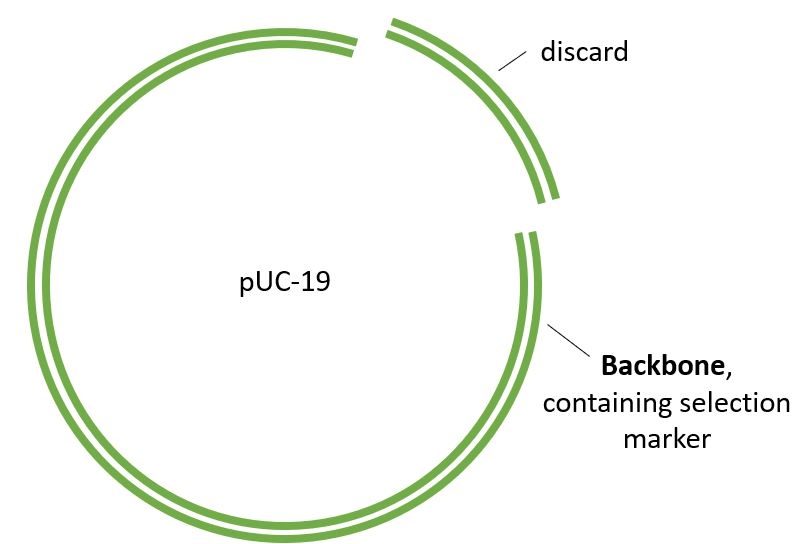
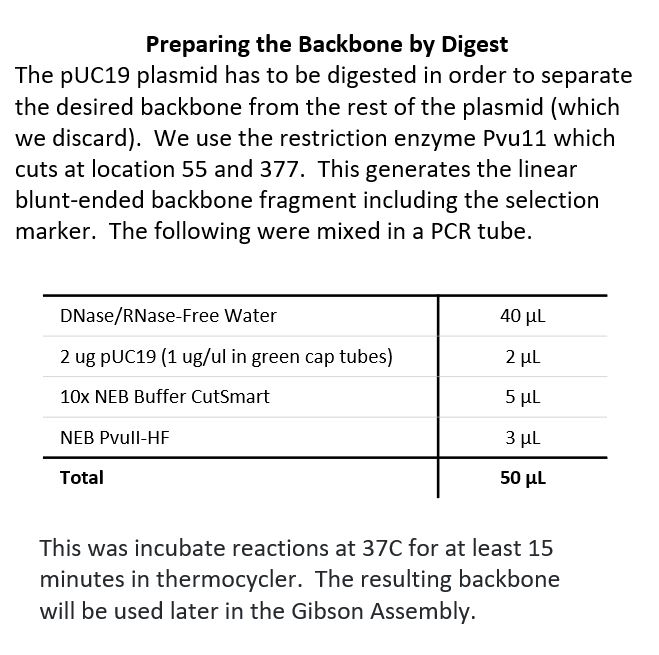
Restriction Digest The pUC-19 plasmid will provide the backbone. The Puv11 restruction enzyme was used to make the appropriate cut sites.
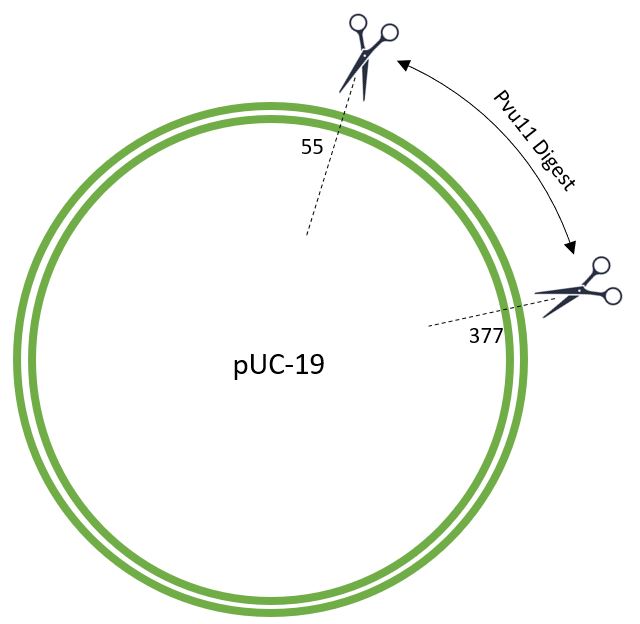
The Backbone is ready to accept the amilCP gene, plus promoter, terminator and RBS.
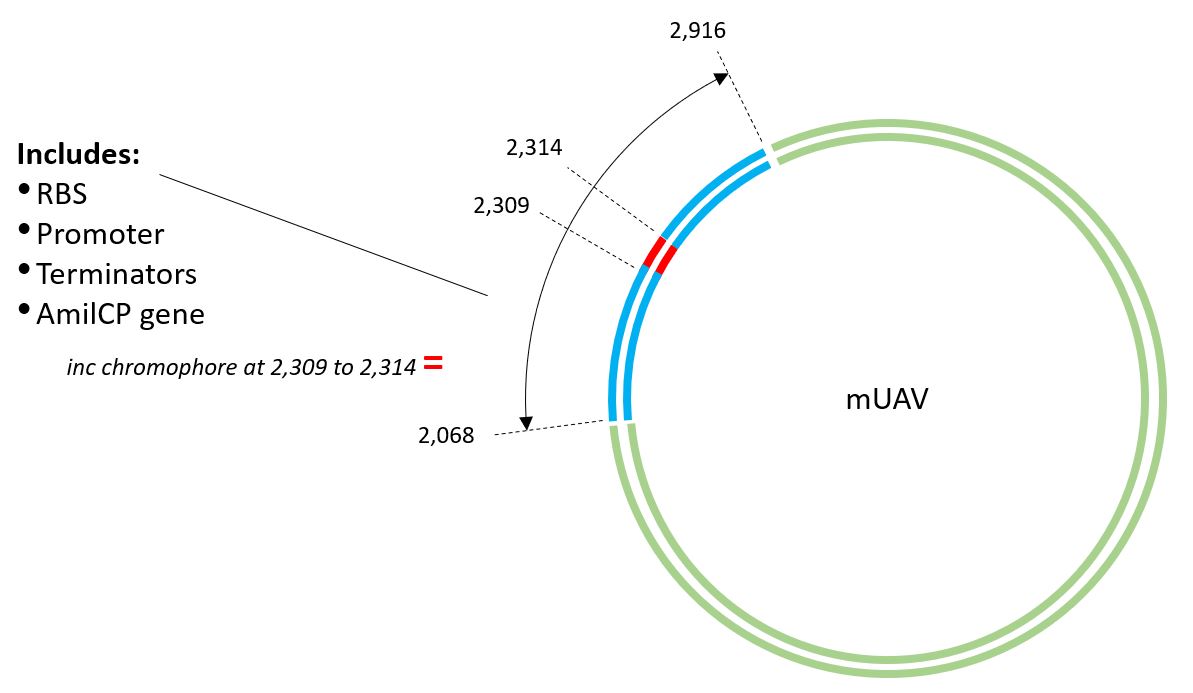
The sequence contains the locations of the key parts of the mUAV plasmid which we want but unfortunaley, the NCBI sequence and Benchling sequence seem to be different by 15Bp. Therefore, I had to do some translation.

This allows to understand the sequence of the mUAV Plasmid and the location of the key entities in the DNA including the AmilCP Gene and its 6Bp Chromophore.
And the AmilCP gene is ready to transfer into the plasmid, but first Primers have to be designed.
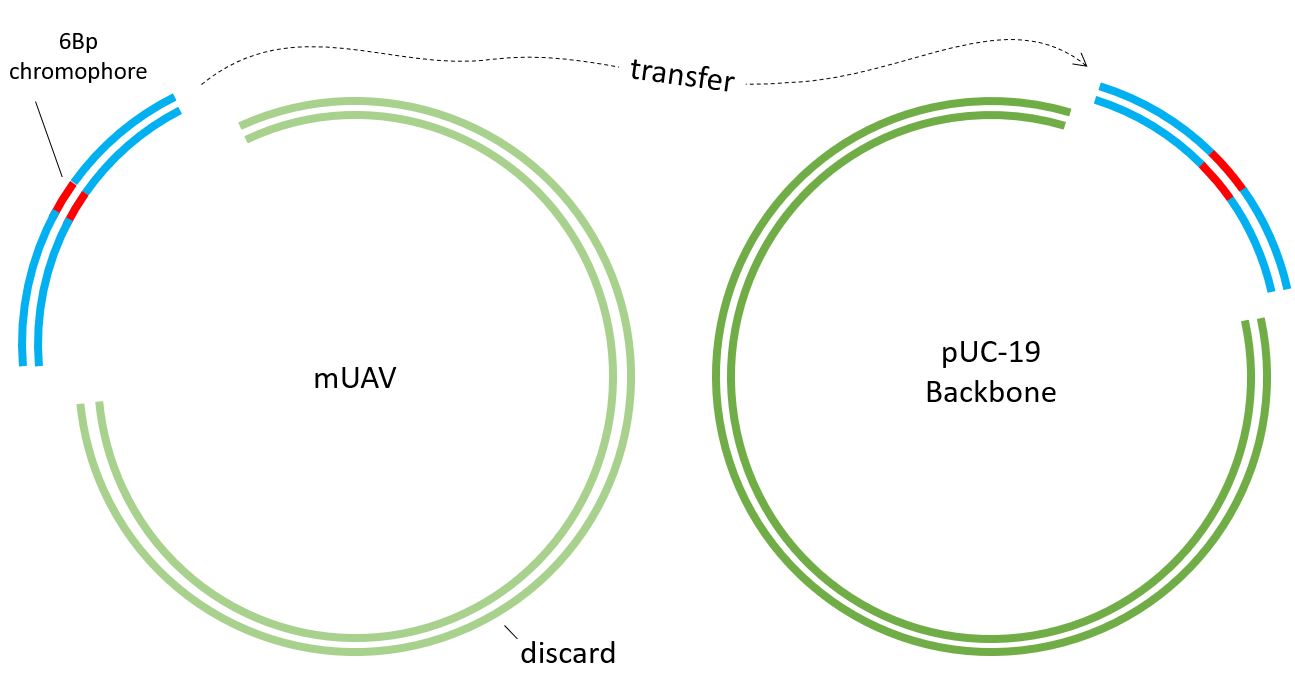
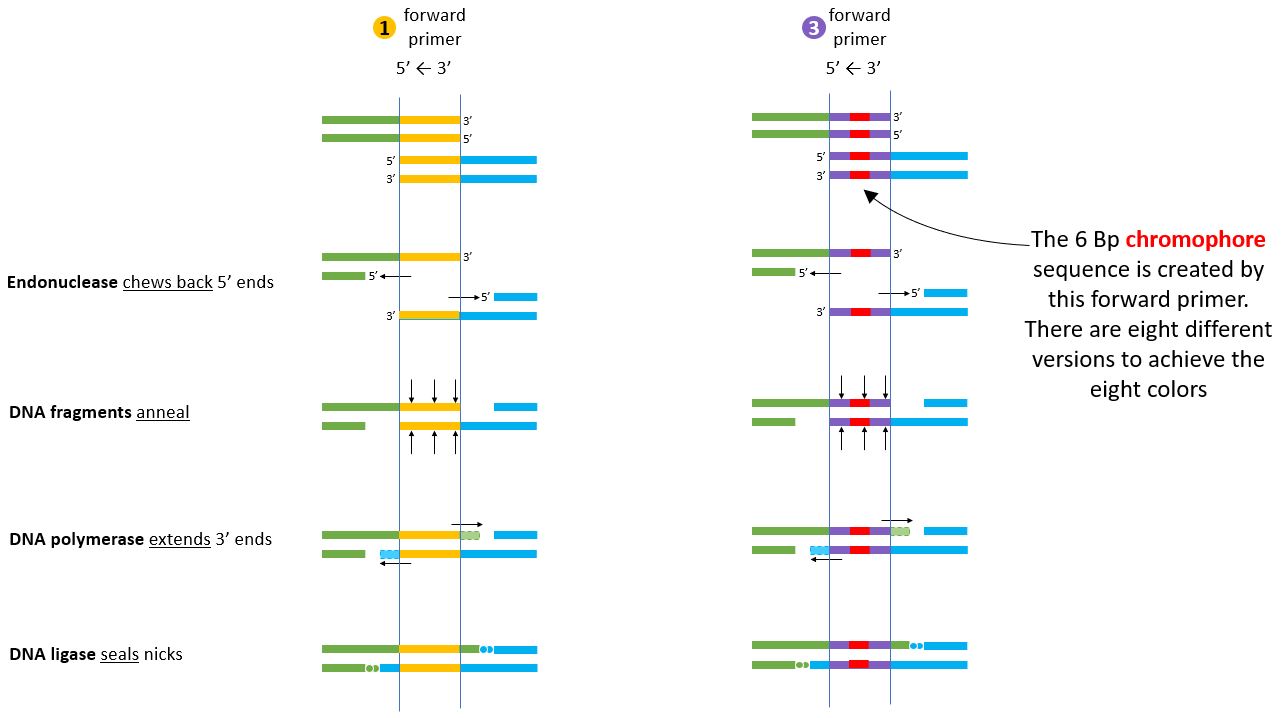
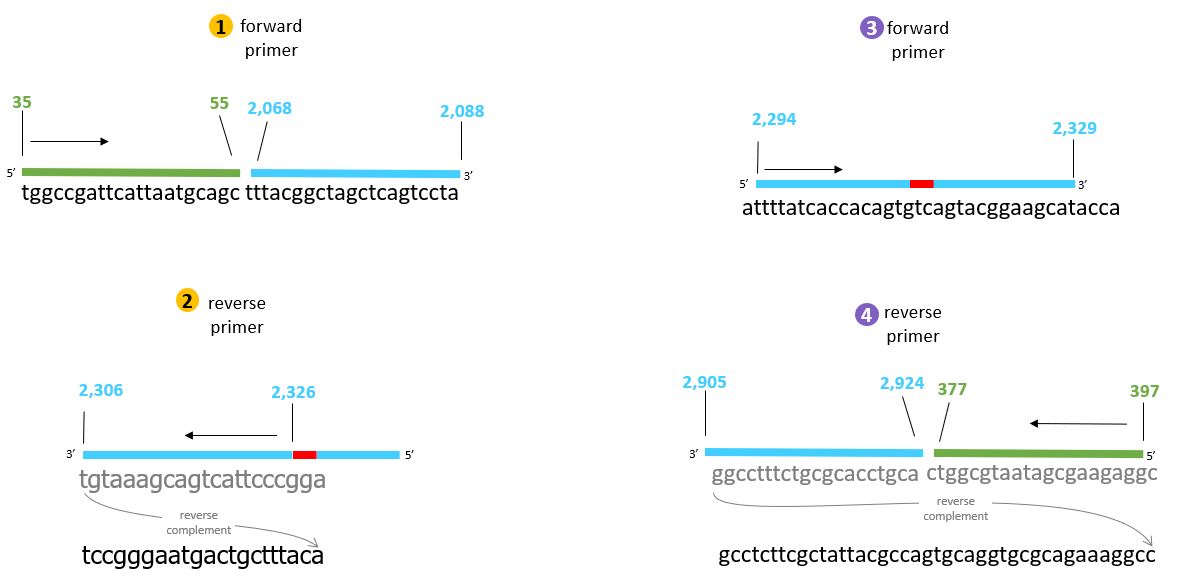
Two sets of primers have to be designed. #3. Foreward Primer is the important one which contains the chromophore sequence. In our case, we cange change the 6Bp sequence to generate 8 different colors.
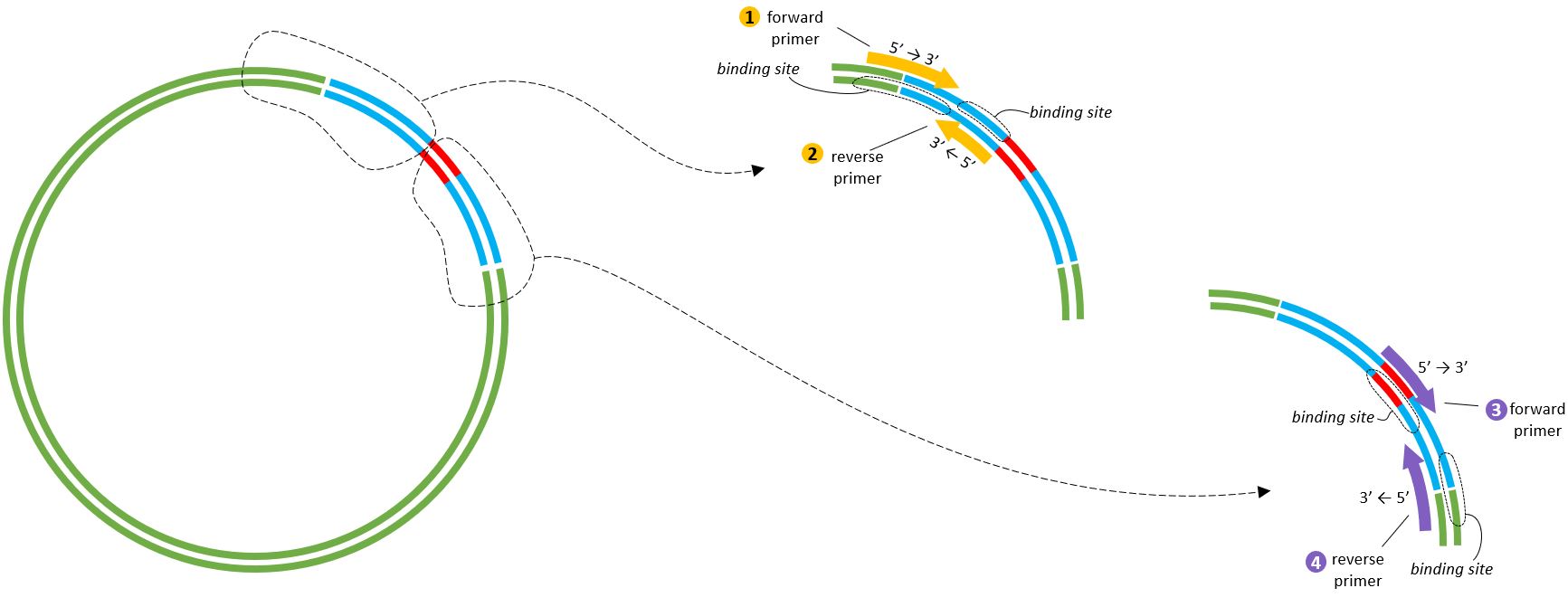
These are the locations of the primers.
But what do the primers do? This sequence shows how the primers are able to create overhangs, on the DNA strands which have to be joined...
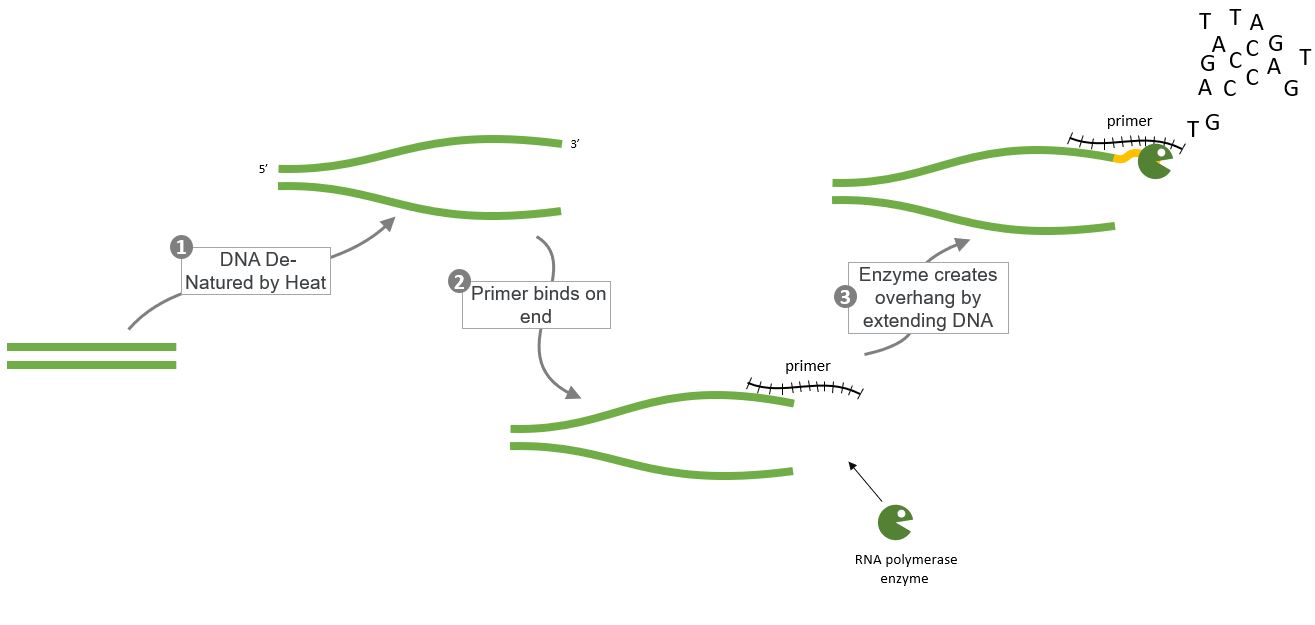
and these overhangs are important because they are used in the Gibson Assembly Process.
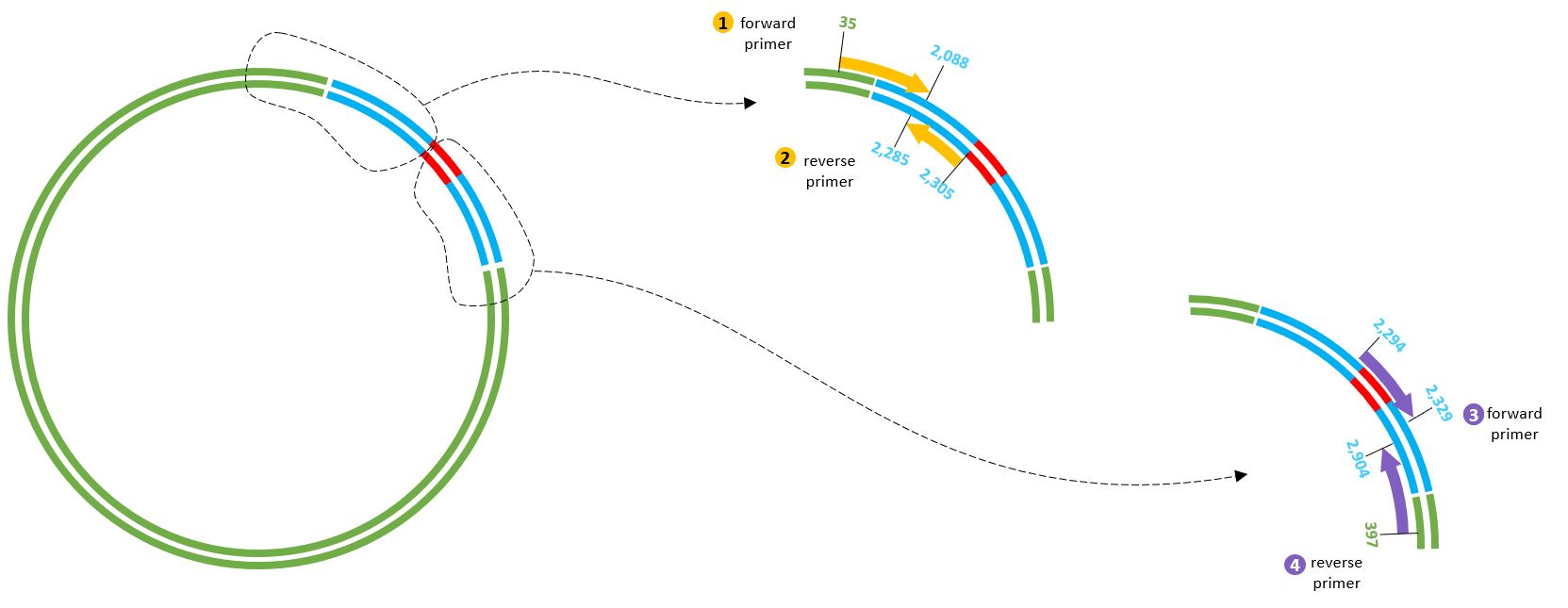
The specific primers are shown here...
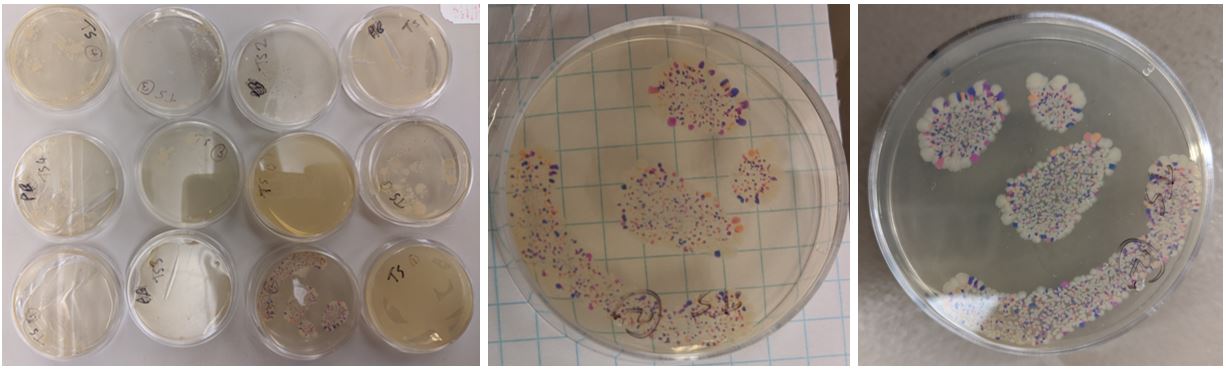
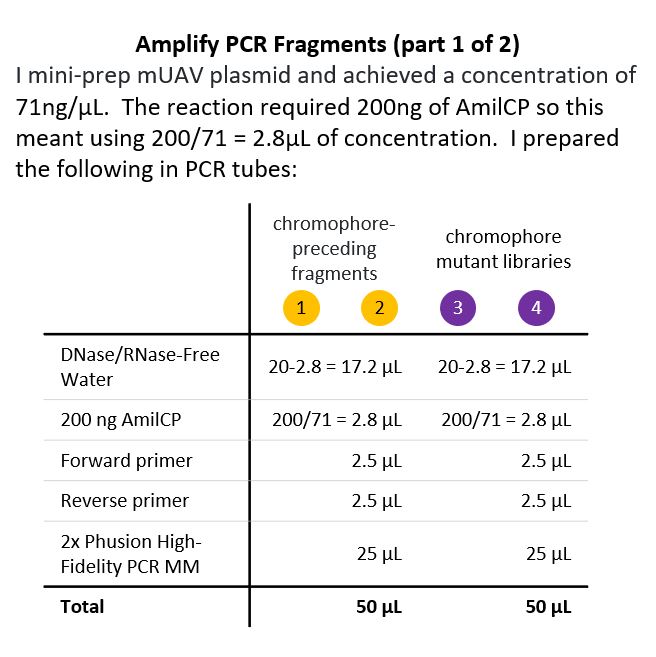
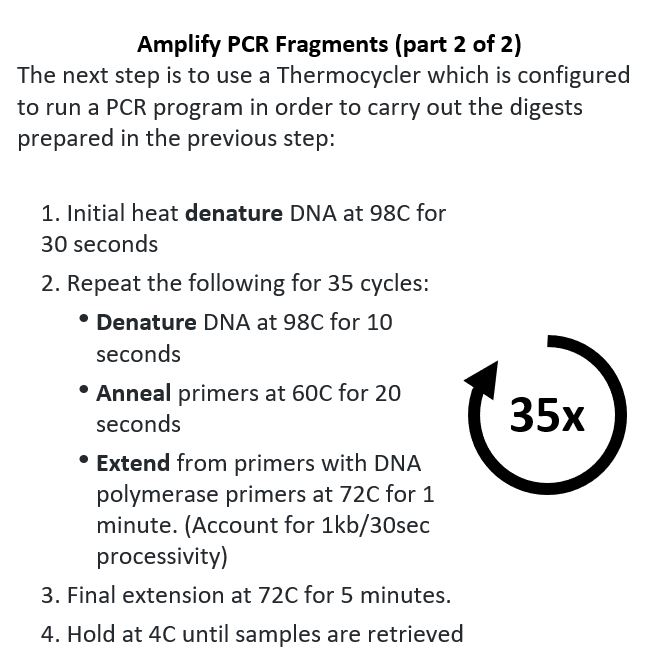
The Lab Part! Now that I had designed the primers, I had to do the lab work



Final Result is that some plates worked, but some were beautifully successful: