Bio Design
Isolate DNA from E.coli, run enzymatic reactions, show the changes and produce a DNA ladder
Software: use snapgene to read the sequence files
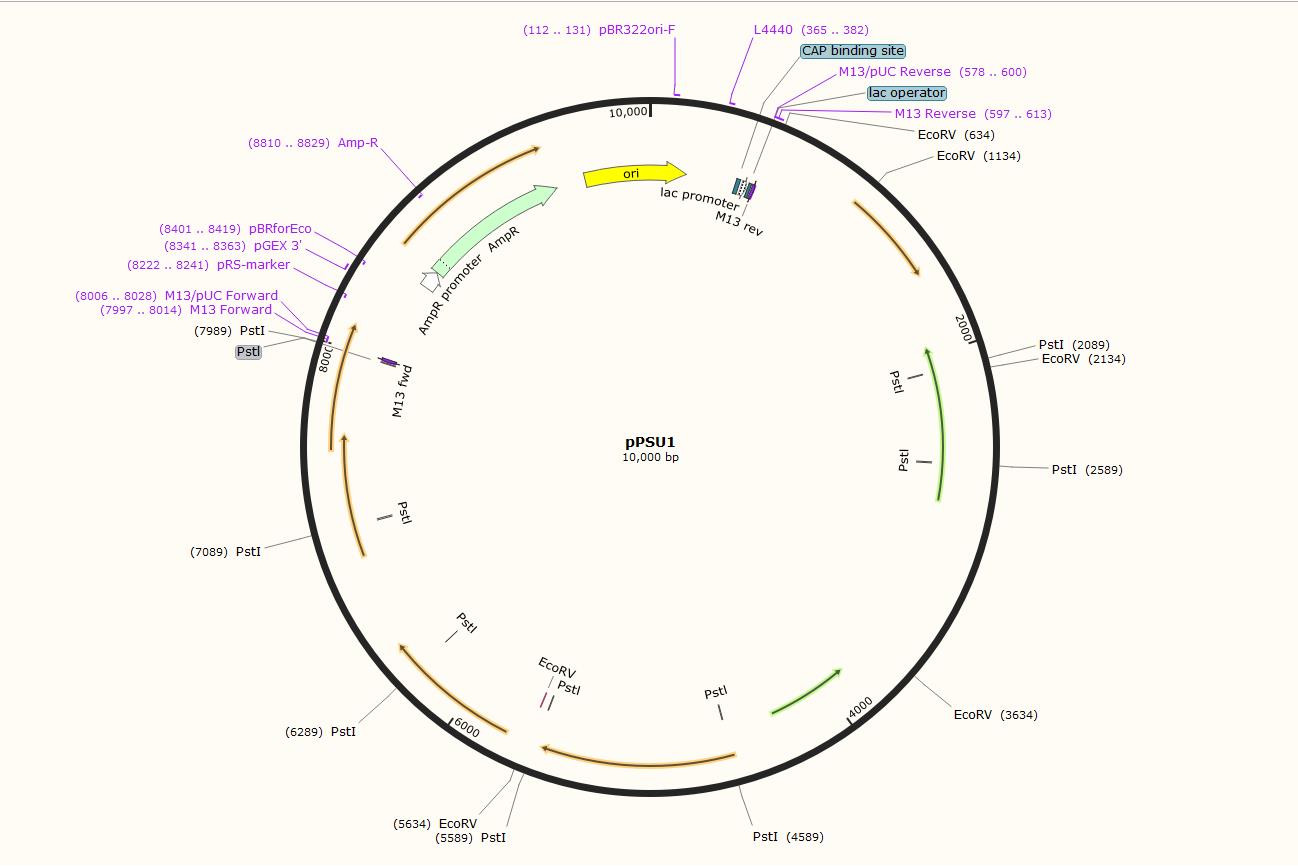
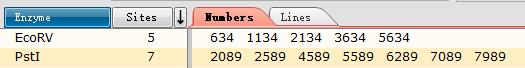
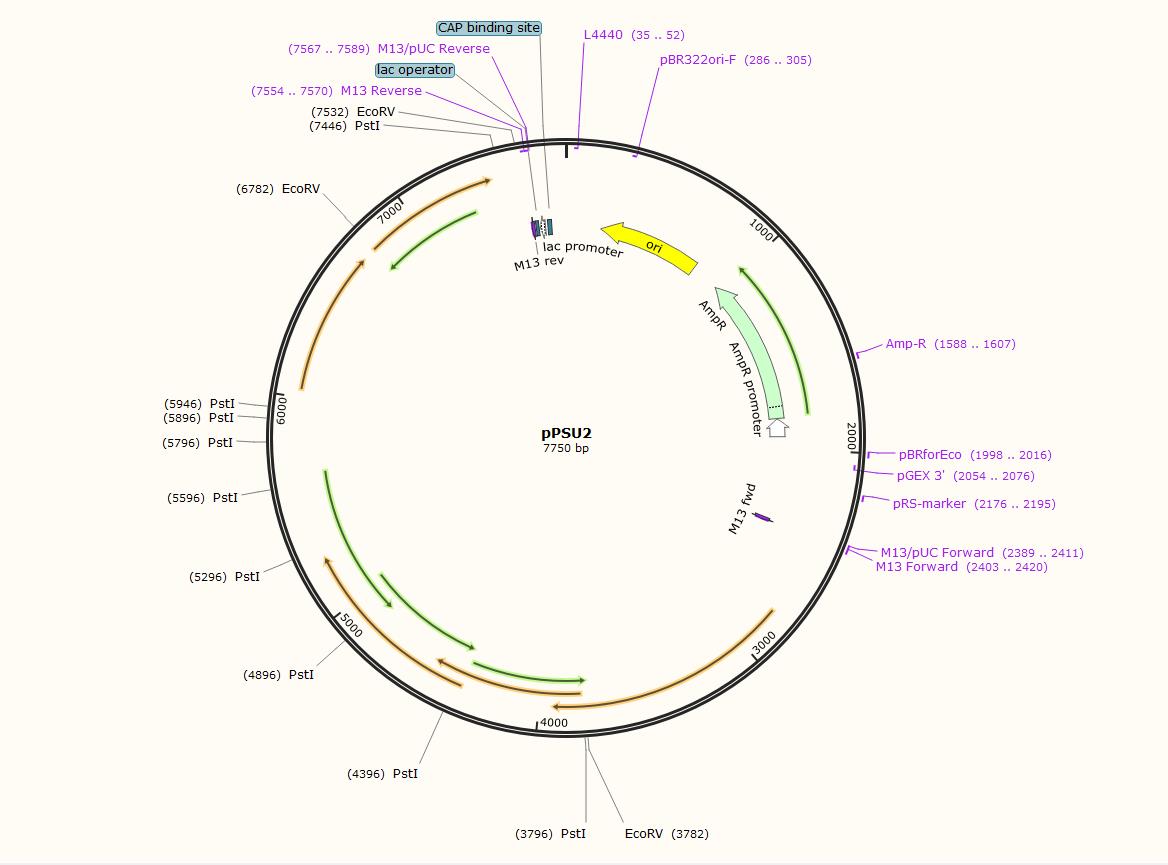
The distance between the Pstl (CTGCAG) sites and EcoRV (GATATC) sites shows as below

Harvest pPSU1 and pPSU2 DNA from overnight E. coli culture using the ZymoPure Plasmid Miniprep Kit and elute with water.

Measure the concentration of your pPSU1 and pPSU2 DNA preps using the Nanodrop spectrophotometer.

The concentration of pPSU1 DNA I yielded is 1893.4ng/ul
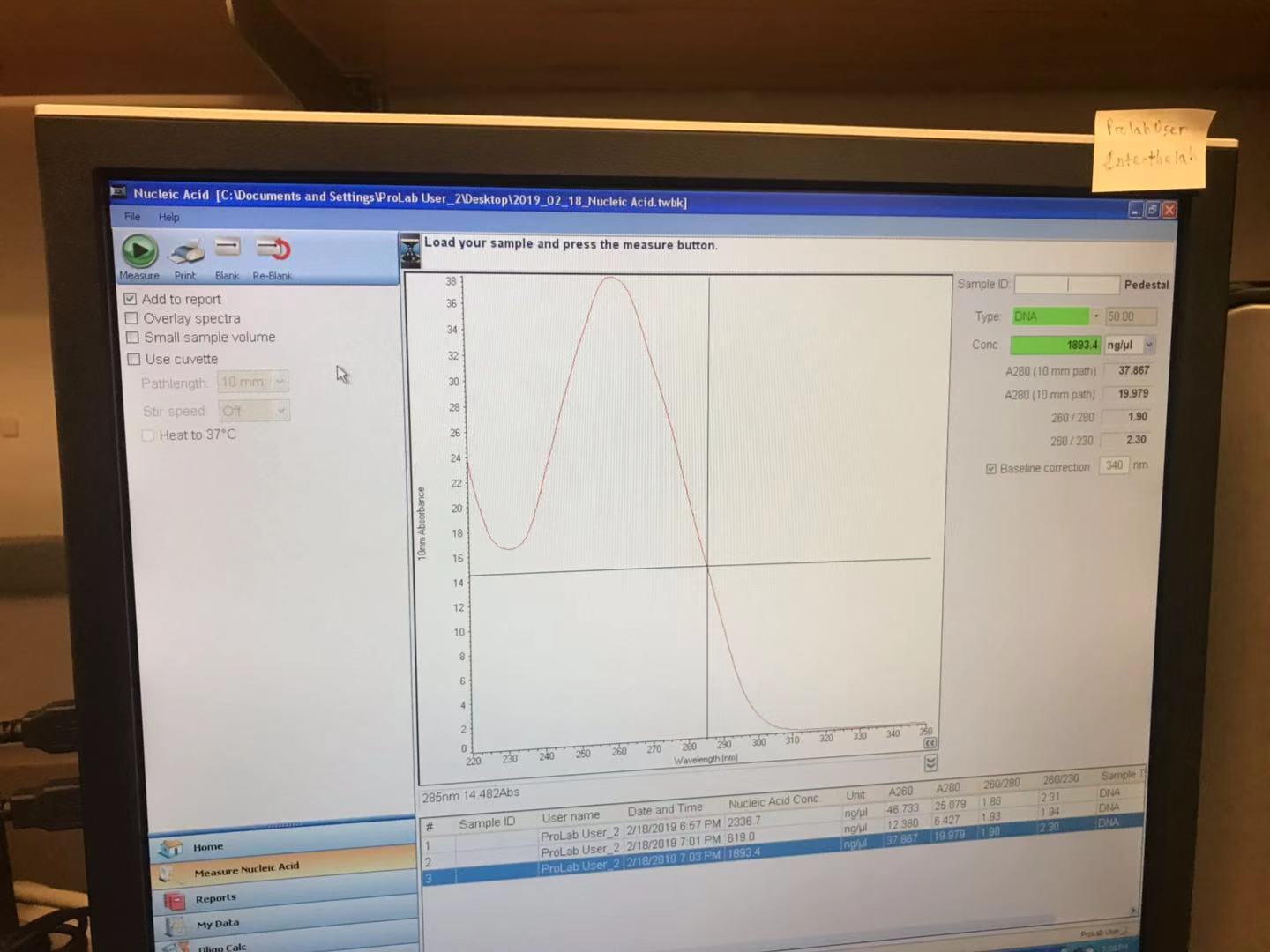
The concentration of pPSU2 DNA I yielded is 1375.1ng/ul
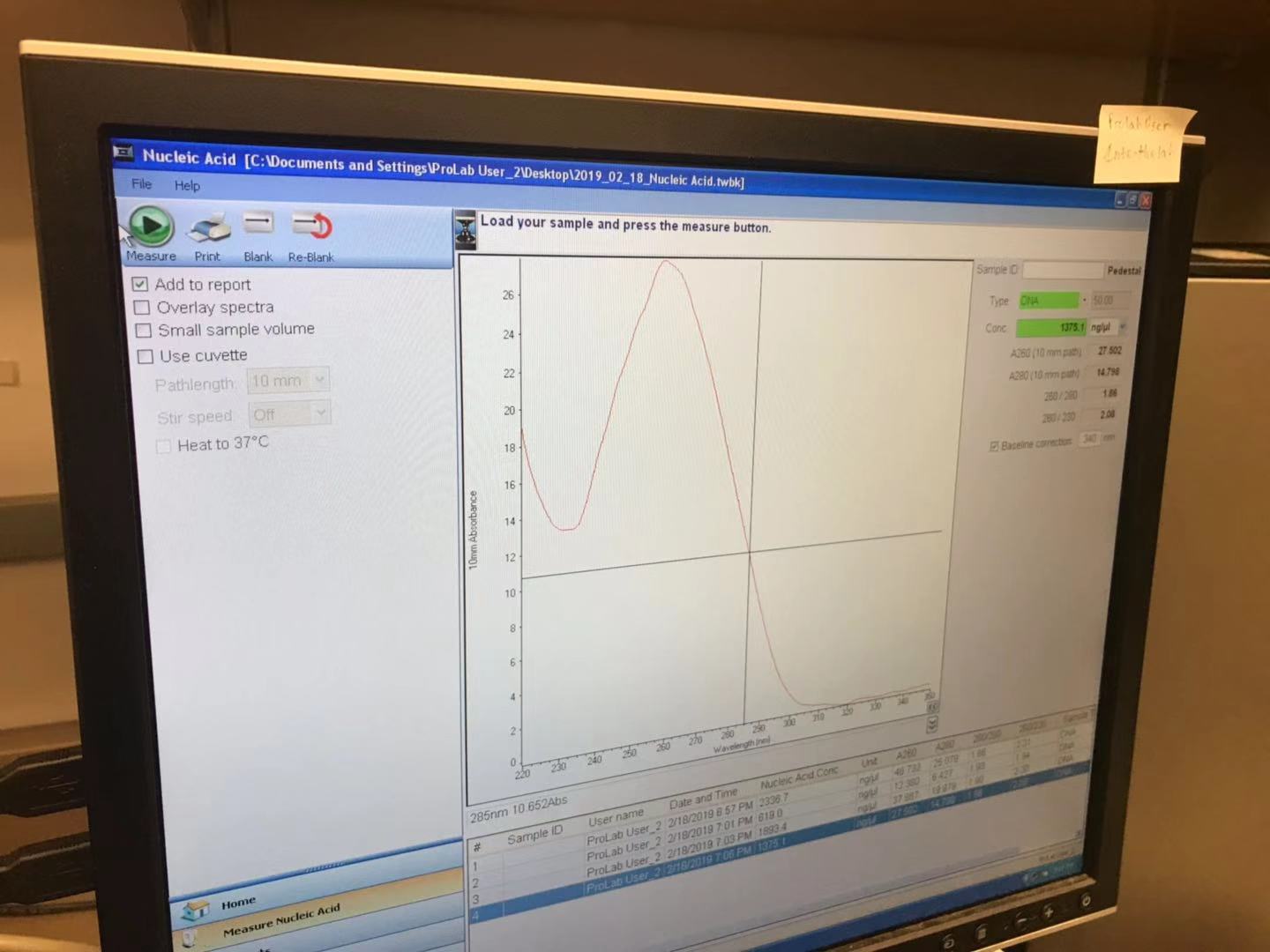
Prepare reactions with EcoRV-HF & Pstl-HF:

Incubate at 37C in thermocycler :

The concentration of pPSU1 DNA is 1893.4ng/ul so I used 4ul of it and added 71.72ul water to dilute the concentration into 100ng/ul
The concentration of pPSU2 DNA is 1375.1ng/ul so I used 4ul of it and added 51ul water to dilute the concentration into 100ng/ul
Making DNA Ladders
Use: 50ul each
NEB 100 bp Reference Ladder
NEB 1 kb Reference Ladder
Uncut pPSU1 Neg Control
Uncut pPSU2 Neg Control
EcoRV-HF Digest Reaction Solution
PstI-HF Digest Reaction Solution

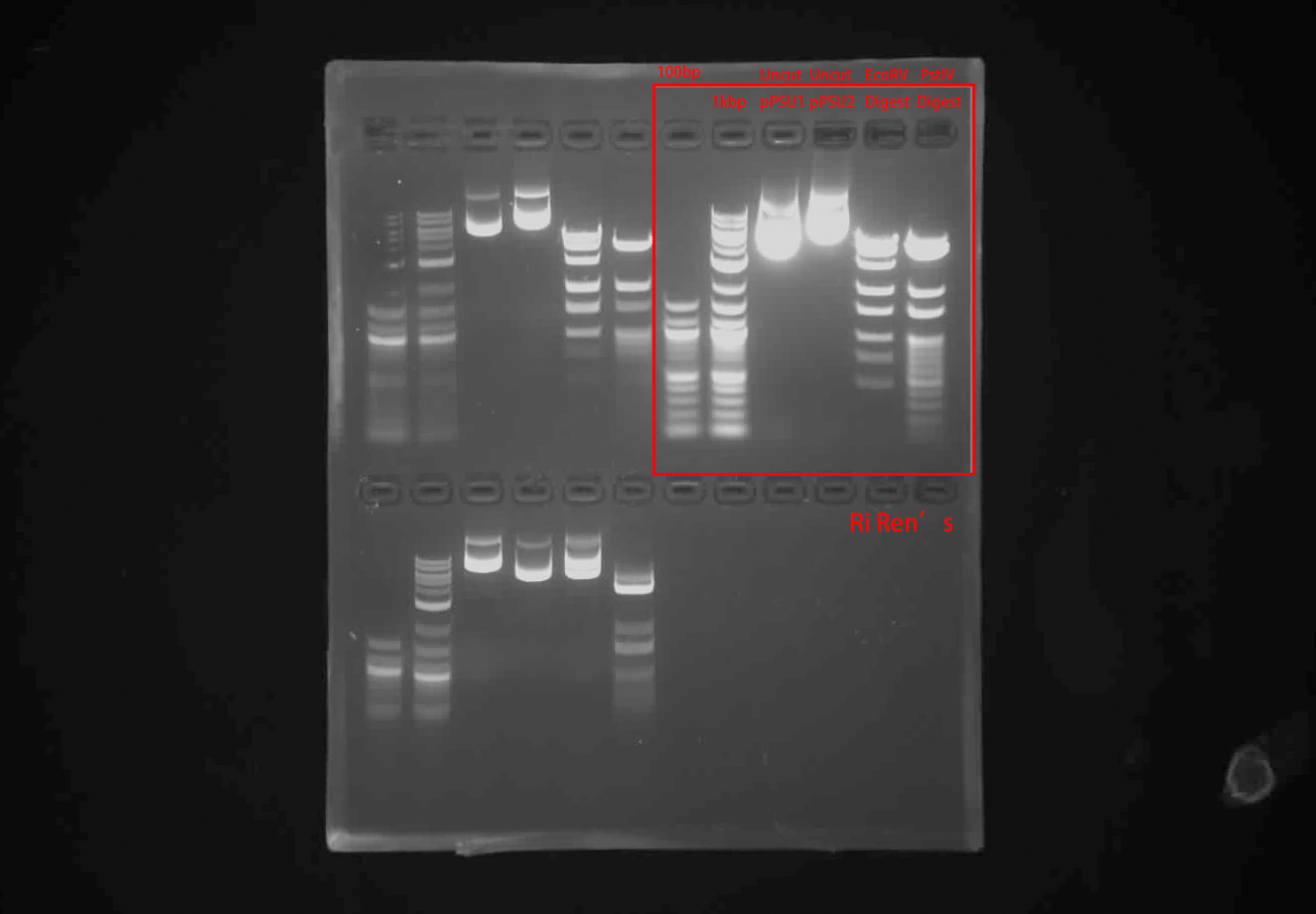
Run the reactions on a 1% agarose gel stained with SYBR-Safe


Image the gels with transilluminator


DNA Ladder image yielded