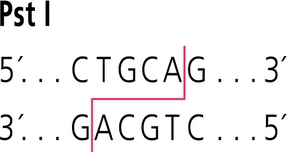
Lab notes:
Following the procedure we collected the E. coli culture and buffers from the ZymoPure kit.
We took the E. coli samples that had the plasmids in them, suspended them, lysed them, neutralized and then washed with buffers.
For our first lab we created DNA ladders using pPSU1 and pPSU2 using the ZymoPure Plasmid Miniprep Kit. The Lab procudure can be found here.
plasmid: Small, typically circular, strand of DNA
restriction enzyme: Enzyme that cuts "cleaves" DNA at a specific sequence of bases.
sticky ends: Unpaired nuceotides at the end of a DNA strant. Also called "cohesive ends" and "overhangs". These sticky ends are useful because they ensure the cut DNA is in the correct direction when inserted into a plasmid.
supernatant: Liquid that separates from solid residue
copy number: Refers to the number of replications a plasmid will have in a host cell. Plasmids will be identified as low, mediumm or high copy number. Plasmids regulate themselves to ensure they do no overly burden the host cell.

Following the procedure we collected the E. coli culture and buffers from the ZymoPure kit.
We took the E. coli samples that had the plasmids in them, suspended them, lysed them, neutralized and then washed with buffers.




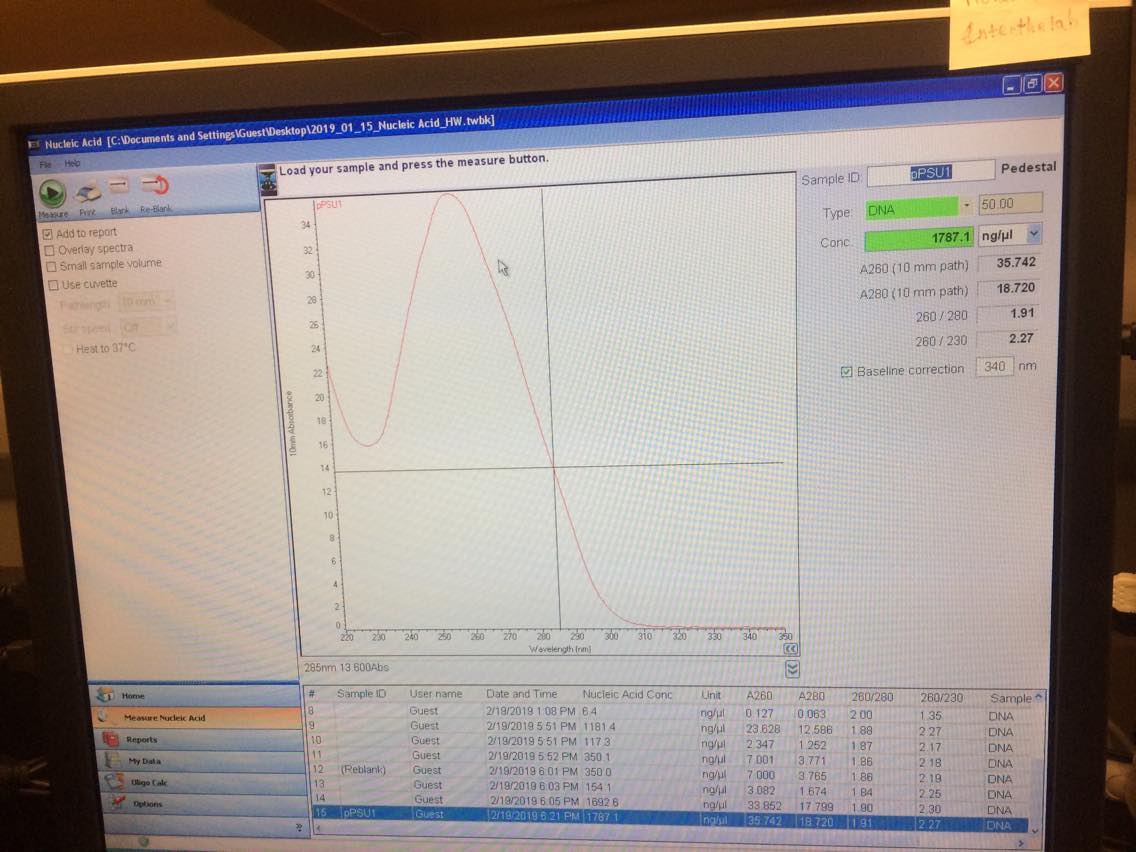
| pPSU1 | pPSU2 | |
| Initial concentration [ng/uL] | 1787 | 1814 |
| Diluted and remeasured [ng/uL] | 243 | 367 |
| Amount needed for 1ug of DNA | 4.1 uL | 2.7 uL |

Q. What software are you using to read the sequence files?
A. Benchling
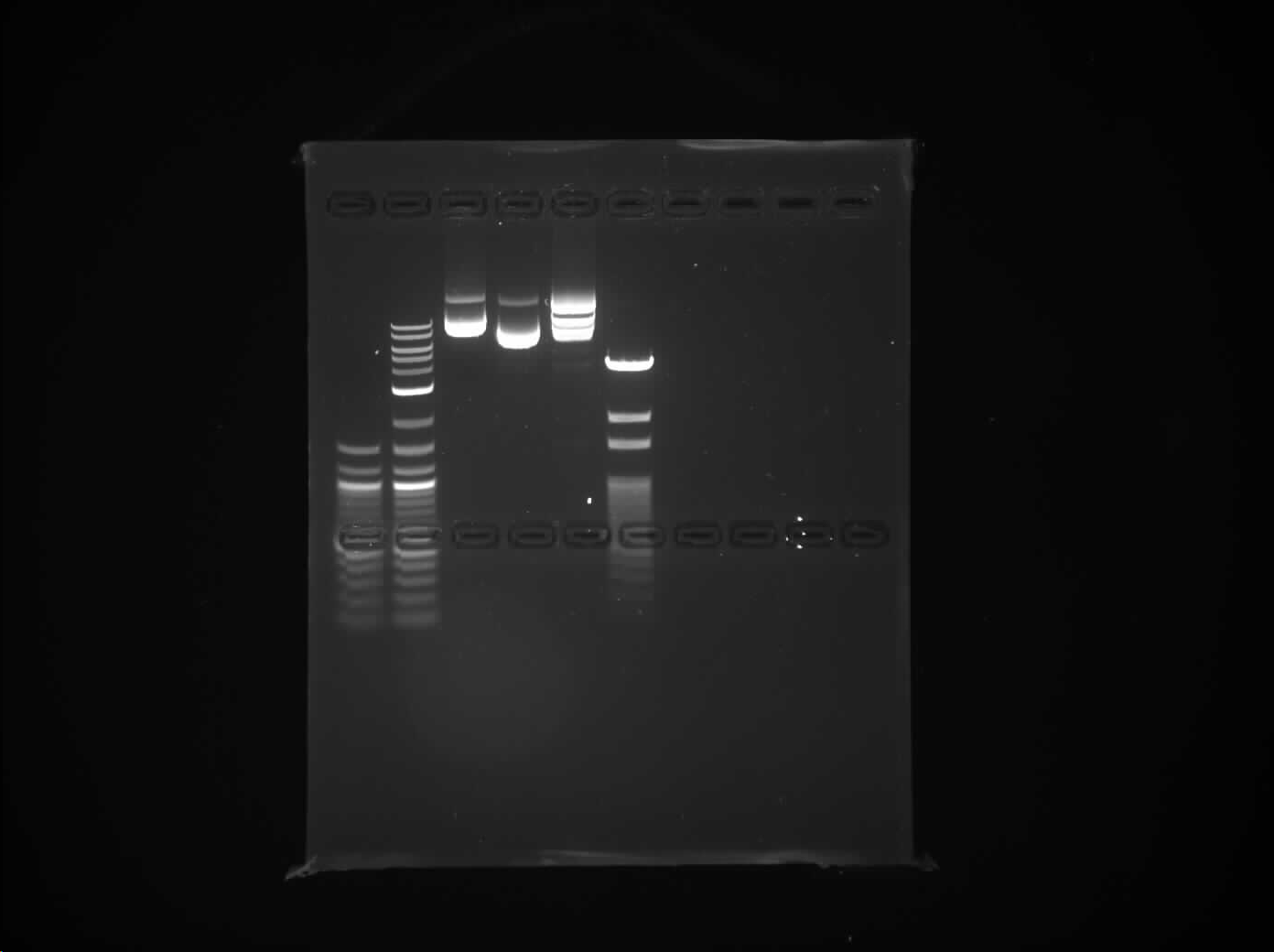
Q. What are the distances between the PstI (CTGCAG) sites in each plasmid? What are the distances between the EcoRV (GATATC) sites in each plasmid?
A. pPSU1 has 7 Pstl (CTGCAG) cleavage sites and EcoRV has 5 (GATATC) cleavage sites.
Distance between cleavage sites in pPSU1 by Pstl: 494, 1994, 994, 694, 794, 894, 4094 [basepairs]
Distance between cleavage sites in pPSU1 by EcoRV:
pPSU2 has 9 Pstl (CTGCAG) cleavage sites and EcoRV has 3 (GATATC) cleavage sites.
Distance between cleavage sites in pPSU2 by Pstl: 1494, 44, 94, 194, 294, 394, 494, 594, 4094
Distance between cleavage sites in pPSU2 by EcoRV: 744, 2994, 3994
These can be found by highlighting the distance between the genes on benching.
Q. Where do PstI and EcoRV cut within their recognition sequences?
A. The sticky ends created by Pst1 and EcoRV can be seen below. These were copied from Sigma-Aldrich