Vocabulary:
Transformation: "Transformation is the process that occurs when a cell ingests foreign DNA from its surroundings."
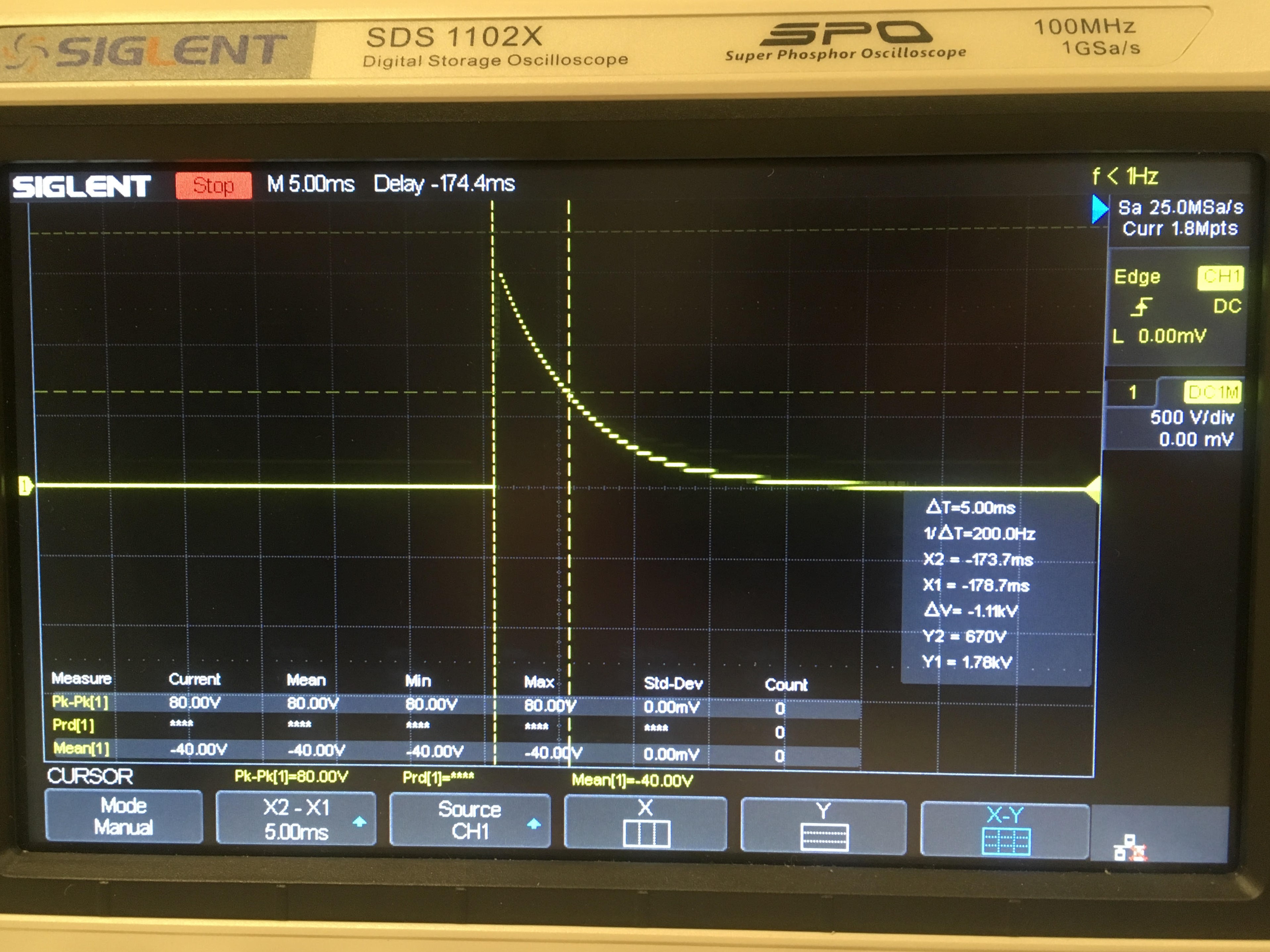
electroporation: "The action or process of introducing DNA or chromosomes into bacteria or other cells using a pulse of electricity to briefly open the pores in the cell membranes."
DIY version of an electroporator
Heat shock"Heat shock transformation uses a calcium rich environment provided by calcium chloride to counteract the electrostatic repulsion between the plasmid DNA and bacterial cellular membrane. A sudden increase in temperature creates pores in the plasma membrane of the bacteria and allows for plasmid DNA to enter the bacterial cell."
cuvette: "a straight-sided, optically clear container for holding liquid samples in a spectrophotometer or other instrument." We used a cuvette for electroporation.
glass beads: Used to spead a bacteria cultures evenly on a petri dish.
Enzyme: "A substance produced by a living organism which acts as a catalyst to bring about a specific biochemical reaction."
ROP protein: (repressor of primer). a protein that regulates replication.
kissing complex: "Kissing" is refereing to RNA that folds back on itself and binds to a section of opposite bases. Two kissing stem loops can also interact, called loop-loop pseudoknots. These interaction are responsible for forming tertiary or quaternary structure of many RNAs.
Origin of replication: a particular sequence in a genome at which replication is initiated.
competent cells: a cell that is capable of taking up exogenous DNA from its surrounding environment.